6OVT
 
 | | Crystal Structure of IlvD from Mycobacterium tuberculosis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dihydroxy-acid dehydratase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Almo, S.C, Grove, T.L, Bonanno, J.B, Baker, E.N, Bashiri, G. | | Deposit date: | 2019-05-08 | | Release date: | 2019-08-07 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The active site of theMycobacterium tuberculosisbranched-chain amino acid biosynthesis enzyme dihydroxyacid dehydratase contains a 2Fe-2S cluster.
J.Biol.Chem., 294, 2019
|
|
6P78
 
 | | queuine lyase from Clostridium spiroforme bound to SAM and queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5S)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-1,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, IRON/SULFUR CLUSTER, Queuine lyase, ... | | Authors: | Almo, S.C, Grove, T.L. | | Deposit date: | 2019-06-05 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.726 Å) | | Cite: | Discovery of novel bacterial queuine salvage enzymes and pathways in human pathogens.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1AAM
 
 | | THE STRUCTURAL BASIS FOR THE ALTERED SUBSTRATE SPECIFICITY OF THE R292D ACTIVE SITE MUTANT OF ASPARTATE AMINOTRANSFERASE FROM E. COLI | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Almo, S.C, Smith, D.L, Danishefsky, A.T, Ringe, D. | | Deposit date: | 1993-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis for the altered substrate specificity of the R292D active site mutant of aspartate aminotransferase from E. coli.
Protein Eng., 7, 1994
|
|
1AAW
 
 | | THE STRUCTURAL BASIS FOR THE ALTERED SUBSTRATE SPECIFICITY OF THE R292D ACTIVE SITE MUTANT OF ASPARTATE AMINOTRANSFERASE FROM E. COLI | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Almo, S.C, Smith, D.L, Danishefsky, A.T, Ringe, D. | | Deposit date: | 1993-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis for the altered substrate specificity of the R292D active site mutant of aspartate aminotransferase from E. coli.
Protein Eng., 7, 1994
|
|
1HQZ
 
 | | Cofilin homology domain of a yeast actin-binding protein ABP1P | | Descriptor: | ACTIN-BINDING PROTEIN | | Authors: | Strokopytov, B.V, Fedorov, A.A, Mahoney, N, Drubin, D.G, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-12-20 | | Release date: | 2001-12-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phased translation function revisited: structure solution of the cofilin-homology domain from yeast actin-binding protein 1 using six-dimensional searches.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7RBW
 
 | | Structure of Biliverdin-binding Serpin of Boana punctata (polka-dot tree frog) | | Descriptor: | BILIVERDINE IX ALPHA, Biliverdin bindin serpin | | Authors: | Fedorov, E, Manoilov, K.Y, Verkhusha, V, Almo, S.C, Ghosh, A. | | Deposit date: | 2021-07-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Functional Characterization of a Biliverdin-Binding Near-Infrared Fluorescent Protein From the Serpin Superfamily.
J.Mol.Biol., 434, 2021
|
|
1QPV
 
 | | YEAST COFILIN | | Descriptor: | YEAST COFILIN | | Authors: | Fedorov, A.A, Lappalainen, P, Fedorov, E.V, Drubin, D.G, Almo, S.C. | | Deposit date: | 1999-05-29 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure determination of yeast cofilin.
Nat.Struct.Biol., 4, 1997
|
|
5WHY
 
 | | Structural Insights into Thioether Bond Formation in the Biosynthesis of Sactipeptides | | Descriptor: | CALCIUM ION, IRON/SULFUR CLUSTER, Radical SAM domain protein, ... | | Authors: | Grove, T.L, Himes, P, Bowers, A, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2017-07-18 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Structural Insights into Thioether Bond Formation in the Biosynthesis of Sactipeptides.
J. Am. Chem. Soc., 139, 2017
|
|
5IZZ
 
 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157, Triple Surface Mutant K158A_K223A_K313A) in complex with metahydroxyphenylacetate, thermal exchange of ligand | | Descriptor: | 3-HYDROXYPHENYLACETATE, DI(HYDROXYETHYL)ETHER, TRAP TRANSPORTER SOLUTE BINDING PROTEIN | | Authors: | Vetting, M.W, Al Obaidi, N.F, Hogle, S.L, Dupont, C.L, Almo, S.C. | | Deposit date: | 2016-03-26 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157, Triple Surface Mutant K158A_K223A_K313A) in complex with metahydroxyphenylacetate, thermal exchange of ligand
To be published
|
|
6X6P
 
 | | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Herrera, N.G, Morano, N.C, Celikgil, A, Georgiev, G.I, Malonis, R, Lee, J.H, Tong, K, Vergnolle, O, Massimi, A, Yen, L.Y, Noble, A.J, Kopylov, M, Bonanno, J.B, Garrett-Thompson, S.C, Hayes, D.B, Brenowitz, M, Garforth, S.J, Eng, E.T, Lai, J.R, Almo, S.C. | | Deposit date: | 2020-05-28 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis.
Biorxiv, 2020
|
|
2AB6
 
 | |
8GAC
 
 | | Crystal structure of a high affinity CTLA-4 binder | | Descriptor: | 1,2-ETHANEDIOL, CTLA-4 binder | | Authors: | Yang, W, Almo, S.C, Baker, D, Ghosh, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of High Affinity Binders to Convex Protein Target Sites.
Biorxiv, 2024
|
|
8GAD
 
 | |
8GAB
 
 | | Crystal structure of CTLA-4 in complex with a high affinity CTLA-4 binder | | Descriptor: | CTLA-4 binder, Cytotoxic T-lymphocyte protein 4, POTASSIUM ION | | Authors: | Yang, W, Almo, S.C, Baker, D, Ghosh, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Design of High Affinity Binders to Convex Protein Target Sites.
Biorxiv, 2024
|
|
5L19
 
 | | Crystal Structure of a human FasL mutant | | Descriptor: | SULFATE ION, Tumor necrosis factor ligand superfamily member 6, ZINC ION | | Authors: | Liu, W, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2016-07-28 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Complex of Human FasL and Its Decoy Receptor DcR3.
Structure, 24, 2016
|
|
5KZK
 
 | | Crystal Structure of rRNA methyltransferase from Sinorhizobium meliloti | | Descriptor: | COBALT (II) ION, Probable RNA methyltransferase, TrmH family, ... | | Authors: | Dey, D, Hegde, R.P, Almo, S.C, Ramakumar, S, Ramagopal, U.A, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-07-25 | | Release date: | 2017-08-02 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of rRNA methyltransferase from Sinorhizobium meliloti
To Be Published
|
|
5L0Z
 
 | | Crystal Structure of AdoMet bound rRNA methyltransferase from Sinorhizobium meliloti | | Descriptor: | COBALT (II) ION, Probable RNA methyltransferase, TrmH family, ... | | Authors: | Dey, D, Hegde, R.P, Almo, S.C, Ramakumar, S, Ramagopal, U.A. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of AdoMet bound rRNA methyltransferase from Sinorhizobium meliloti
To Be Published
|
|
5L36
 
 | |
6NG3
 
 | | Crystal structure of human CD160 and HVEM complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD160 antigen,Tumor necrosis factor receptor superfamily member 14, MAGNESIUM ION, ... | | Authors: | Liu, W, Bonanno, J, Almo, S.C. | | Deposit date: | 2018-12-21 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Basis of CD160:HVEM Recognition.
Structure, 27, 2019
|
|
6NG9
 
 | |
6NGG
 
 | |
6OI0
 
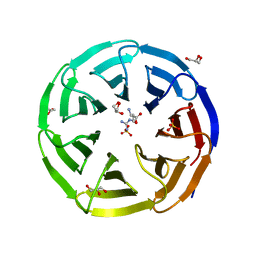 | | Crystal structure of human WDR5 in complex with L-arginine | | Descriptor: | ARGININE, GLYCEROL, SULFATE ION, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
1ZVM
 
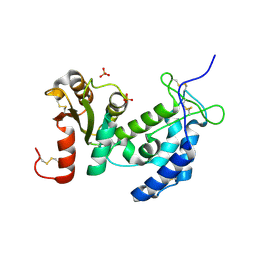 | | Crystal structure of human CD38: cyclic-ADP-ribosyl synthetase/NAD+ glycohydrolase | | Descriptor: | ADP-ribosyl cyclase 1, SULFATE ION | | Authors: | Shi, W, Yang, T, Almo, S.C, Schramm, V.L, Sauve, A. | | Deposit date: | 2005-06-02 | | Release date: | 2006-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human CD38: Cyclic-ADP-ribosyl synthetase/NAD+ glycohydrolase
To be Published
|
|
2A2L
 
 | |
2ACG
 
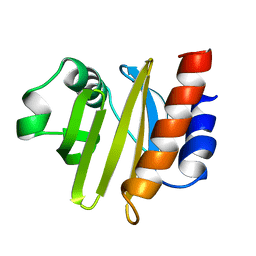 | | ACANTHAMOEBA CASTELLANII PROFILIN II | | Descriptor: | PROFILIN II | | Authors: | Fedorov, A.A, Magnus, K.A, Graupe, M.H, Lattman, E.E, Pollard, T.D, Almo, S.C. | | Deposit date: | 1994-08-30 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structures of isoforms of the actin-binding protein profilin that differ in their affinity for phosphatidylinositol phosphates.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
