1COF
 
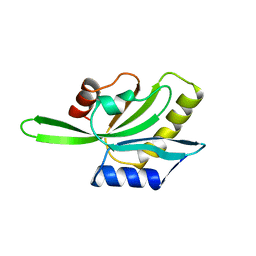 | | YEAST COFILIN, ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | COFILIN | | Authors: | Fedorov, A.A, Lappalainen, P, Fedorov, E.V, Drubin, D.G, Almo, S.C. | | Deposit date: | 1996-11-26 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure determination of yeast cofilin.
Nat.Struct.Biol., 4, 1997
|
|
1D4X
 
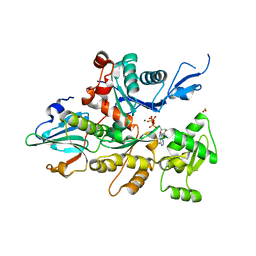 | | Crystal Structure of Caenorhabditis Elegans Mg-ATP Actin Complexed with Human Gelsolin Segment 1 at 1.75 A resolution. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, C. ELEGANS ACTIN 1/3, CALCIUM ION, ... | | Authors: | Vorobiev, S, Ono, S, Almo, S.C. | | Deposit date: | 1999-10-06 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of nonvertebrate actin: implications for the ATP hydrolytic mechanism.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
3NQD
 
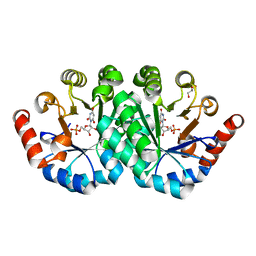 | | Crystal structure of the mutant I96T of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-06-29 | | Release date: | 2011-05-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
1DQN
 
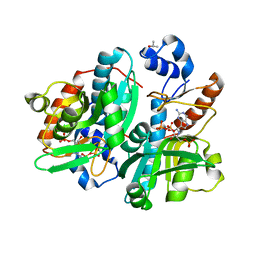 | | CRYSTAL STRUCTURE OF GIARDIA GUANINE PHOSPHORIBOSYLTRANSFERASE COMPLEXED WITH A TRANSITION STATE ANALOGUE | | Descriptor: | GUANINE PHOSPHORIBOSYLTRANSFERASE, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Shi, W, Munagala, N.R, Wang, C.C, Li, C.M, Tyler, P.C, Furneaux, R.H, Grubmeyer, C, Schramm, V.L, Almo, S.C. | | Deposit date: | 2000-01-04 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Giardia lamblia guanine phosphoribosyltransferase at 1.75 A(,).
Biochemistry, 39, 2000
|
|
5YTI
 
 | | Crystal structure of flagellar hook associated protein-3 (HAP-3: Q5ZW61_LEGPH) from Legionella pneumophila | | Descriptor: | CADMIUM ION, Flagellar hook associated protein type 3 FlgL | | Authors: | Lankipalli, S, Hegde, R.P, Dey, D, Almo, S.C, Ramagopal, U.A. | | Deposit date: | 2017-11-18 | | Release date: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of hook associated protein-3 (HAP-3: Q5ZW61_LEGPH) from Legionella pneumophila
To be published
|
|
3PBY
 
 | | Crystal structure of the mutant L123S of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, GLYCEROL, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
3PBW
 
 | | Crystal structure of the mutant L123N of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
3PBU
 
 | | Crystal structure of the mutant I96S of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
3PC0
 
 | | Crystal structure of the mutant V155S of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, GLYCEROL, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
3PBV
 
 | | Crystal structure of the mutant I96T of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, GLYCEROL, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
1CJF
 
 | | PROFILIN BINDS PROLINE-RICH LIGANDS IN TWO DISTINCT AMIDE BACKBONE ORIENTATIONS | | Descriptor: | 7-HYDROXY-4-METHYL-3-(2-HYDROXY-ETHYL)COUMARIN, PROTEIN (HUMAN PLATELET PROFILIN), PROTEIN (PROLINE PEPTIDE) | | Authors: | Mahoney, N.M, Fedorov, A.A, Fedorov, E, Rozwarski, D.A, Almo, S.C. | | Deposit date: | 1999-04-13 | | Release date: | 1999-07-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Profilin binds proline-rich ligands in two distinct amide backbone orientations.
Nat.Struct.Biol., 6, 1999
|
|
1CQA
 
 | | BIRCH POLLEN PROFILIN | | Descriptor: | PROFILIN | | Authors: | Fedorov, A.A, Ball, T, Mahoney, N.M, Valenta, R, Almo, S.C. | | Deposit date: | 1996-07-26 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular basis for allergen cross-reactivity: crystal structure and IgE-epitope mapping of birch pollen profilin.
Structure, 5, 1997
|
|
1DQT
 
 | | THE CRYSTAL STRUCTURE OF MURINE CTLA4 (CD152) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CYTOTOXIC T LYMPHOCYTE ASSOCIATED ANTIGEN 4 | | Authors: | Ostrov, D.A, Shi, W, Schwartz, J.C, Almo, S.C, Nathenson, S.G. | | Deposit date: | 2000-01-05 | | Release date: | 2000-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of murine CTLA-4 and its role in modulating T cell responsiveness.
Science, 290, 2000
|
|
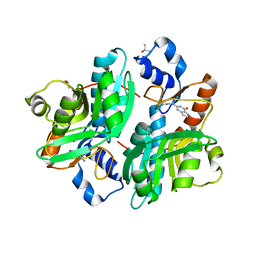
1G2P
 
 | | CRYSTAL STRUCTURE OF ADENINE PHOSPHORIBOSYLTRANSFERASE | | Descriptor: | ADENINE PHOSPHORIBOSYLTRANSFERASE 1, SULFATE ION | | Authors: | Shi, W, Tanaka, K.S.E, Almo, S.C, Schramm, V.L. | | Deposit date: | 2000-10-20 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of adenine phosphoribosyltransferase from Saccharomyces cerevisiae.
Biochemistry, 40, 2001
|
|
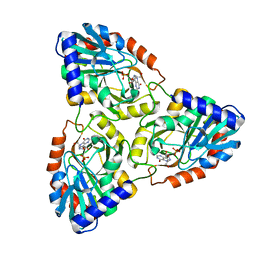
1G2Q
 
 | |
1DQP
 
 | | CRYSTAL STRUCTURE OF GIARDIA GUANINE PHOSPHORIBOSYLTRANSFERASE COMPLEXED WITH IMMUCILLING | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-1-(S)-(9-DEAZAGUANIN-9-YL)-D-RIBITOL, GUANINE PHOSPHORIBOSYLTRANSFERASE, ISOPROPYL ALCOHOL | | Authors: | Shi, W, Munagala, N.R, Wang, C.C, Li, C.M, Tyler, P.C, Furneaux, R.H, Grubmeyer, C, Schramm, V.L, Almo, S.C. | | Deposit date: | 2000-01-04 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Giardia lamblia guanine phosphoribosyltransferase at 1.75 A(,).
Biochemistry, 39, 2000
|
|
1G2O
 
 | | CRYSTAL STRUCTURE OF PURINE NUCLEOSIDE PHOSPHORYLASE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH A TRANSITION-STATE INHIBITOR | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PHOSPHATE ION, PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Shi, W, Basso, L.A, Tyler, P.C, Furneaux, R.H, Blanchard, J.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2000-10-20 | | Release date: | 2001-08-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of purine nucleoside phosphorylase from Mycobacterium tuberculosis in complexes with immucillin-H and its pieces.
Biochemistry, 40, 2001
|
|
6AQU
 
 | | Crystal Structure of Plasmodium falciparum purine nucleoside phosphorylase: The M183L mutant | | Descriptor: | PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Harijan, R.K, Ducati, R.G, Namanja-Magliano, H.A, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-08-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Genetic resistance to purine nucleoside phosphorylase inhibition inPlasmodium falciparum.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6AQS
 
 | | Crystal structure of Plasmodium falciparum purine nucleoside phosphorylase (V181D) mutant complexed with DADMe-ImmG and phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, ... | | Authors: | Harijan, R.K, Ducati, R.G, Namanja-Magliano, H.A, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-08-21 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Genetic resistance to purine nucleoside phosphorylase inhibition in
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6AYR
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with butylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
1EZR
 
 | | CRYSTAL STRUCTURE OF NUCLEOSIDE HYDROLASE FROM LEISHMANIA MAJOR | | Descriptor: | CALCIUM ION, NUCLEOSIDE HYDROLASE | | Authors: | Shi, W, Schramm, V.L, Almo, S.C. | | Deposit date: | 2000-05-11 | | Release date: | 2000-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nucleoside hydrolase from Leishmania major. Cloning, expression, catalytic properties, transition state inhibitors, and the 2.5-a crystal structure.
J.Biol.Chem., 274, 1999
|
|
6AYQ
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with methylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, GLYCEROL | | Authors: | Cameron, S.A, Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
6AYT
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with pyrazinylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(pyrazin-2-ylsulfanyl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
1CFY
 
 | | YEAST COFILIN, MONOCLINIC CRYSTAL FORM | | Descriptor: | COFILIN | | Authors: | Fedorov, A.A, Lappalainen, P, Fedorov, E.V, Drubin, D.G, Almo, S.C. | | Deposit date: | 1996-11-26 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure determination of yeast cofilin.
Nat.Struct.Biol., 4, 1997
|
|
1DFC
 
 | | CRYSTAL STRUCTURE OF HUMAN FASCIN, AN ACTIN-CROSSLINKING PROTEIN | | Descriptor: | FASCIN | | Authors: | Fedorov, A.A, Fedorov, E.V, Ono, S, Matsumura, F, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 1999-11-18 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure, evolutionary conservation, and conformational dynamics of Homo sapiens fascin-1, an F-actin crosslinking protein.
J.Mol.Biol., 400, 2010
|
|
