8OX9
 
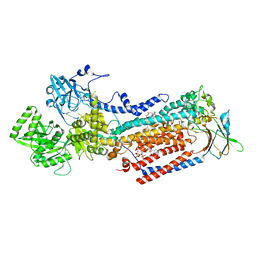 | | Cryo-EM structure of ATP8B1-CDC50A in E2P active conformation with bound PC | | Descriptor: | (2R)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dioctanoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dieudonne, T, Kummerer, F, Juknaviciute Laursen, M, Stock, C, Kock Flygaard, R, Khalid, S, Lenoir, G, Lyons, J.A, Lindorff-Larsen, K, Nissen, P. | | Deposit date: | 2023-05-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Activation and substrate specificity of the human P4-ATPase ATP8B1.
Nat Commun, 14, 2023
|
|
4JBX
 
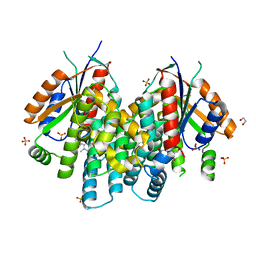 | | Crystal structure of thymidine kinase from Herpes simplex virus type 1 in complex with SK-78 | | Descriptor: | 1,2-ETHANEDIOL, 6-[3-hydroxy-2-(hydroxymethyl)prop-1-en-1-yl]-1,5-dimethylpyrimidine-2,4(1H,3H)-dione, SULFATE ION, ... | | Authors: | Pernot, L, Novakovic, I, Westermaier, Y, Perozzo, R, Raic-Malic, S, Scapozza, L. | | Deposit date: | 2013-02-20 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New thymine derivatives as HSV1-TK ligand for the development of PET imaging tracer
To be Published
|
|
1PR1
 
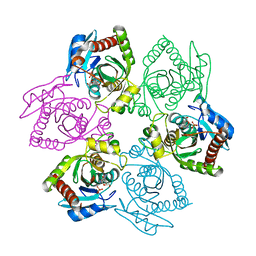 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with Formycin B and Phosphate/Sulfate | | Descriptor: | FORMYCIN B, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
8QOE
 
 | | Inward-facing conformation of the ABC transporter BmrA | | Descriptor: | Multidrug resistance ABC transporter ATP-binding/permease protein BmrA | | Authors: | Di Cesare, M, Kaplan, E, Valimehr, S, Hanssen, E, Orelle, C, Jault, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | The transport activity of the multidrug ABC transporter BmrA does not require a wide separation of the nucleotide-binding domains.
J.Biol.Chem., 300, 2023
|
|
1R9G
 
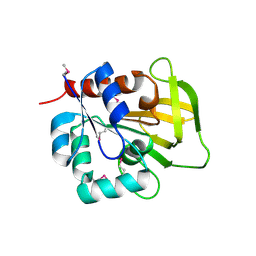 | | Three-dimensional Structure of YaaE from Bacillus subtilis | | Descriptor: | Hypothetical protein yaaE | | Authors: | Bauer, J.A, Bennett, E.M, Begley, T.P, Ealick, S.E. | | Deposit date: | 2003-10-29 | | Release date: | 2004-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional Structure of YaaE from Bacillus subtilis, a Glutaminase Implicated in Pyridoxal-5'-phosphate Biosynthesis.
J.Biol.Chem., 279, 2004
|
|
1YFW
 
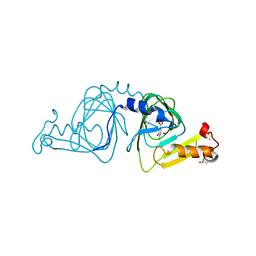 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase from Ralstonia metallidurans complexed with 4-chloro-3-hydroxyanthranilic acid and O2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate-3,4-dioxygenase, 4-CHLORO-3-HYDROXYANTHRANILIC ACID, ... | | Authors: | Zhang, Y, Colabroy, K.L, Begley, T.P, Ealick, S.E. | | Deposit date: | 2005-01-04 | | Release date: | 2005-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Studies on 3-Hydroxyanthranilate-3,4-dioxygenase: The Catalytic Mechanism of a Complex Oxidation Involved in NAD Biosynthesis.
Biochemistry, 44, 2005
|
|
1YAK
 
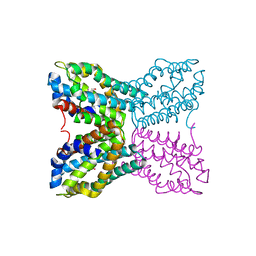 | | Complex of Bacillus subtilis TenA with 4-amino-2-methyl-5-hydroxymethylpyrimidine | | Descriptor: | 4-AMINO-5-HYDROXYMETHYL-2-METHYLPYRIMIDINE, Transcriptional activator tenA | | Authors: | Toms, A.V, Haas, A.L, Park, J.-H, Begley, T.P, Ealick, S.E. | | Deposit date: | 2004-12-17 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural characterization of the regulatory proteins TenA and TenI from Bacillus subtilis and identification of TenA as a thiaminase II.
Biochemistry, 44, 2005
|
|
4JBY
 
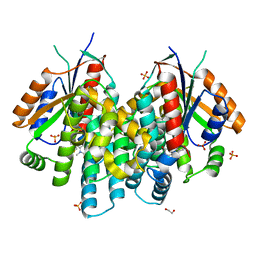 | | Crystal structure of thymidine kinase from Herpes simplex virus type 1 in complex with F-SK78 | | Descriptor: | 1,2-ETHANEDIOL, 6-[(1Z)-3-fluoro-2-(hydroxymethyl)prop-1-en-1-yl]-1,5-dimethylpyrimidine-2,4(1H,3H)-dione, SULFATE ION, ... | | Authors: | Pernot, L, Novakovic, I, Westermaier, Y, Perozzo, R, Raic-Malic, S, Scapozza, L. | | Deposit date: | 2013-02-20 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New thymine derivatives as HSV1-TK ligand for the development of PET imaging tracer
To be Published
|
|
1PR6
 
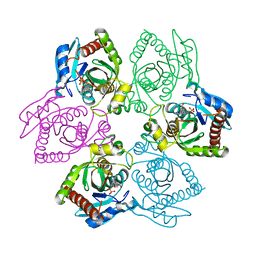 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with 9-beta-D-xylofuranosyladenine and Phosphate/Sulfate | | Descriptor: | 2-(6-AMINO-OCTAHYDRO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
2JMM
 
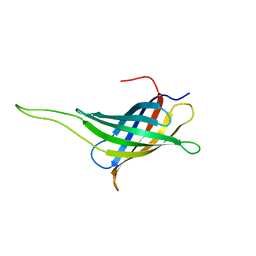 | | NMR solution structure of a minimal transmembrane beta-barrel platform protein | | Descriptor: | Outer membrane protein A | | Authors: | Johansson, M.U, Alioth, S, Hu, K, Walser, R, Koebnik, R, Pervushin, K. | | Deposit date: | 2006-11-20 | | Release date: | 2007-07-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | A minimal transmembrane beta-barrel platform protein studied by nuclear magnetic resonance
Biochemistry, 46, 2007
|
|
1PR5
 
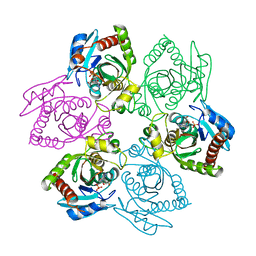 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with 7-deazaadenosine and Phosphate/Sulfate | | Descriptor: | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PR0
 
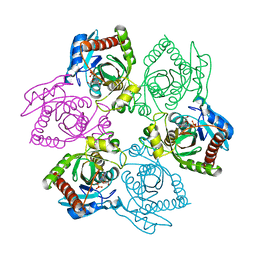 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with Inosine and Phosphate/Sulfate | | Descriptor: | INOSINE, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1TWJ
 
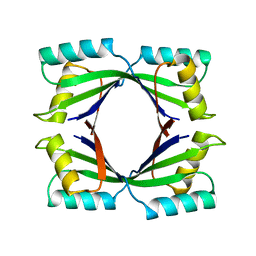 | |
1YFY
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase from Ralstonia metallidurans complexed with 3-hydroxyanthranilic acid | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate-3,4-dioxygenase, ... | | Authors: | Zhang, Y, Colabroy, K.L, Begley, T.P, Ealick, S.E. | | Deposit date: | 2005-01-04 | | Release date: | 2005-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Studies on 3-Hydroxyanthranilate-3,4-dioxygenase: The Catalytic Mechanism of a Complex Oxidation Involved in NAD Biosynthesis.
Biochemistry, 44, 2005
|
|
1YAF
 
 | | Structure of TenA from Bacillus subtilis | | Descriptor: | Transcriptional activator tenA | | Authors: | Toms, A.V, Haas, A.L, Park, J.-H, Begley, T.P, Ealick, S.E. | | Deposit date: | 2004-12-17 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural characterization of the regulatory proteins TenA and TenI from Bacillus subtilis and identification of TenA as a thiaminase II.
Biochemistry, 44, 2005
|
|
2R7K
 
 | | Crystal structure of FAICAR synthetase (PurP) from M. jannaschii complexed with AMPPCP and AICAR | | Descriptor: | 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5'-monophosphate synthetase, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, CHLORIDE ION, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
1PR2
 
 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with 9-beta-D-[2-deoxyribofuranosyl]-6-methylpurine and Phosphate/Sulfate | | Descriptor: | 9-(2-DEOXY-BETA-D-RIBOFURANOSYL)-6-METHYLPURINE, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1U11
 
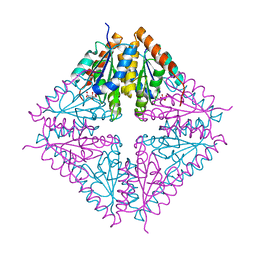 | | PurE (N5-carboxyaminoimidazole Ribonucleotide Mutase) from the acidophile Acetobacter aceti | | Descriptor: | CITRIC ACID, PurE (N5-carboxyaminoimidazole Ribonucleotide Mutase) | | Authors: | Settembre, E.C, Chittuluru, J.R, Mill, C.P, Kappock, T.J, Ealick, S.E. | | Deposit date: | 2004-07-14 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acidophilic adaptations in the structure of Acetobacter aceti N5-carboxyaminoimidazole ribonucleotide mutase (PurE).
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2PMV
 
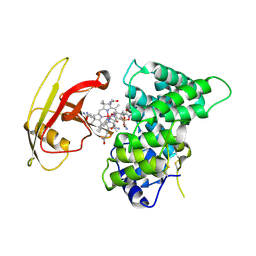 | | Crystal Structure of Human Intrinsic Factor- Cobalamin Complex at 2.6 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COBALAMIN, Gastric intrinsic factor | | Authors: | Mathews, F.S, Gordon, M.M, Chen, Z, Rajashankar, K.R, Ealick, S.E, Alpers, D.H, Sukumar, N. | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human intrinsic factor: Cobalamin complex at 2.6-A resolution
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1SD1
 
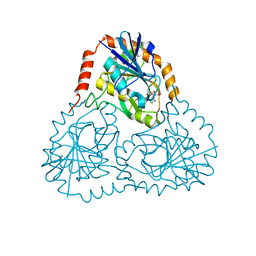 | | STRUCTURE OF HUMAN 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE COMPLEXED WITH FORMYCIN A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, 5'-methylthioadenosine phosphorylase | | Authors: | Lee, J.E, Settembre, E.C, Cornell, K.A, Riscoe, M.K, Sufrin, J.R, Ealick, S.E, Howell, P.L. | | Deposit date: | 2004-02-12 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Comparison of MTA Phosphorylase and MTA/AdoHcy Nucleosidase Explains Substrate Preferences and Identifies Regions Exploitable for Inhibitor Design.
Biochemistry, 43, 2004
|
|
3GMC
 
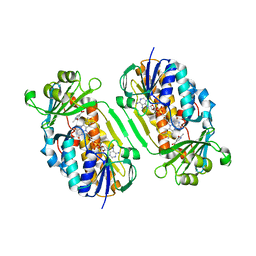 | | Crystal Structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid Oxygenase with substrate bound | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxy-6-methylpyridine-3-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | McCulloch, K.M, Mukherjee, T, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-03-13 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the PLP degradative enzyme 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase from Mesorhizobium loti MAFF303099 and its mechanistic implications.
Biochemistry, 48, 2009
|
|
1ECP
 
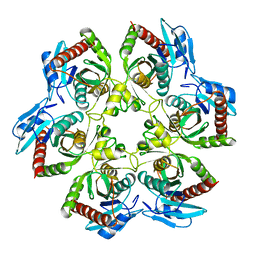 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | Descriptor: | PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Mao, C, Ealick, S.E. | | Deposit date: | 1995-07-13 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Escherichia coli purine nucleoside phosphorylase: a comparison with the human enzyme reveals a conserved topology.
Structure, 5, 1997
|
|
1U1G
 
 | | Structure of E. coli uridine phosphorylase complexed to 5-(m-(benzyloxy)benzyl)barbituric acid (BBBA) | | Descriptor: | 1-((2-HYDROXYETHOXY)METHYL)-5-(3-(BENZYLOXY)BENZYL)-6-HYDROXYPYRIMIDINE-2,4(1H,3H)-DIONE, POTASSIUM ION, Uridine phosphorylase | | Authors: | Bu, W, Settembre, E.C, Ealick, S.E. | | Deposit date: | 2004-07-15 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2R87
 
 | |
1N13
 
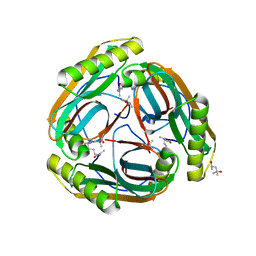 | | The Crystal Structure of Pyruvoyl-dependent Arginine Decarboxylase from Methanococcus jannashii | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, AGMATINE, Pyruvoyl-dependent arginine decarboxylase alpha chain, ... | | Authors: | Tolbert, W.D, Graham, D.E, White, R.H, Ealick, S.E. | | Deposit date: | 2002-10-16 | | Release date: | 2003-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Pyruvoyl-Dependent Arginine Decarboxylase from Methanococcus jannaschii:
Crystal Structures of the Self-Cleaved and S53A Proenzyme Forms
Structure, 11, 2003
|
|
