9G2A
 
 | | Staphylococcus aureus MazF in complex with nanobody 4 | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endoribonuclease MazF, ... | | Authors: | Prolic-Kalinsek, M, Zorzini, V, Haesaerts, S, Loris, R. | | Deposit date: | 2024-07-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.046594 Å) | | Cite: | Staphylococcus aureus MazF in complex with nanobody 4.
To Be Published
|
|
5L6S
 
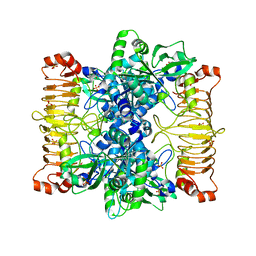 | | Crystal structure of E. coli ADP-glucose pyrophosphorylase (AGPase) in complex with a positive allosteric regulator beta-fructose-1,6-diphosphate (FBP) - AGPase*FBP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Cifuente, J.O, Albesa-Jove, D, Comino, N, Madariaga-Marcos, J, Agirre, J, Lopez-Fernandez, S, Garcia-Alija, M, Guerin, M.E. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural Basis of Glycogen Biosynthesis Regulation in Bacteria.
Structure, 24, 2016
|
|
5L6V
 
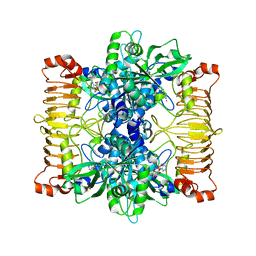 | | Crystal structure of E. coli ADP-glucose pyrophosphorylase (AGPase) in complex with a negative allosteric regulator adenosine monophosphate (AMP) - AGPase*AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glucose-1-phosphate adenylyltransferase, PHOSPHATE ION, ... | | Authors: | Cifuente, J.O, Albesa-Jove, D, Comino, N, Madariaga-Marcos, J, Agirre, J, Lopez-Fernandez, S, Garcia-Alija, M, Guerin, M.E. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.667 Å) | | Cite: | Structural Basis of Glycogen Biosynthesis Regulation in Bacteria.
Structure, 24, 2016
|
|
6TVI
 
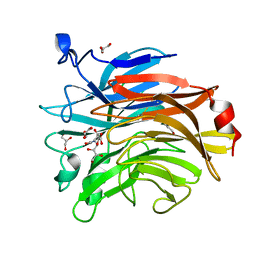 | | Salmonella typhimurium mutant neuraminidase (D100S)+ DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, Sialidase | | Authors: | Garman, E.F, Salinger, M.T, Murray, J.W, Laver, W.G, Kuhn, P, Vimr, E.R. | | Deposit date: | 2020-01-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Salmonella typhimurium mutant neuraminidase (D100S)+ DANA
To Be Published
|
|
9GDM
 
 | |
8G8E
 
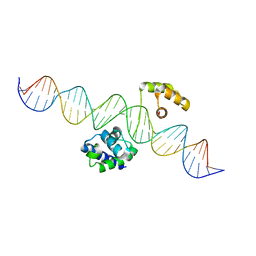 | | Human Oct4 bound to nucleosome with human LIN28B sequence | | Descriptor: | LIN28B DNA (32-MER), POU domain, class 5, ... | | Authors: | Sinha, K.K, Bilokapic, S, Du, Y, Malik, D, Halic, M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Histone modifications regulate pioneer transcription factor cooperativity.
Nature, 619, 2023
|
|
5LYG
 
 | | CRYSTAL STRUCTURE OF INTRACELLULAR B30.2 DOMAIN OF BTN3A1 BOUND TO MALONATE | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 3 member A1, MALONATE ION | | Authors: | Mohammed, F, Baker, A.T, Salim, M, Willcox, B.E. | | Deposit date: | 2016-09-28 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | BTN3A1 Discriminates gamma delta T Cell Phosphoantigens from Nonantigenic Small Molecules via a Conformational Sensor in Its B30.2 Domain.
ACS Chem. Biol., 12, 2017
|
|
5K8Q
 
 | | Crystal Structure of Calcium-loaded Calmodulin in complex with STRA6 CaMBP2-site peptide. | | Descriptor: | CALCIUM ION, Calmodulin, IMIDAZOLE, ... | | Authors: | Stowe, S.D, Clarke, O.B, Cavalier, M.C, Godoy-Ruiz, R, Mancia, F, Weber, D.J. | | Deposit date: | 2016-05-30 | | Release date: | 2016-08-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | Structure of the STRA6 receptor for retinol uptake.
Science, 353, 2016
|
|
7PUK
 
 | | Crystal structure of Endoglycosidase E GH18 domain from Enterococcus faecalis in complex with Man5 product | | Descriptor: | Beta-N-acetylhexosaminidase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Garcia-Alija, M, Du, J.J, Trastoy, B, Sundberg, E.J, Guerin, M. | | Deposit date: | 2021-09-30 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE.
Nat Commun, 13, 2022
|
|
7PUL
 
 | | Crystal structure of Endoglycosidase E GH20 domain from Enterococcus faecalis | | Descriptor: | Beta-N-acetylhexosaminidase, CALCIUM ION, GLY-ALA-GLY-ALA-ALA, ... | | Authors: | Garcia-Alija, M, Du, J.J, Trastoy, B, Sundberg, E.J, Guerin, M. | | Deposit date: | 2021-09-30 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE.
Nat Commun, 13, 2022
|
|
7PUJ
 
 | | Crystal structure of Endoglycosidase E GH18 domain from Enterococcus faecalis | | Descriptor: | Beta-N-acetylhexosaminidase, CHLORIDE ION, ZINC ION | | Authors: | Garcia-Alija, M, Du, J.J, Trastoy, B, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2021-09-30 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE.
Nat Commun, 13, 2022
|
|
5MM8
 
 | |
5LYM
 
 | |
5AD2
 
 | | Bivalent binding to BET bromodomains | | Descriptor: | (3R)-4-(2-{4-[1-(3-chloro[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidinyl]phenoxy}ethyl)-1,3-dimethyl-2-piperazinone, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
5AD3
 
 | | Bivalent binding to BET bromodomains | | Descriptor: | 3-methoxy-N-[2-[4-[1-(3-methoxy-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidyl]phenoxy]ethyl]-N-methyl-[1,2,4]triazolo[4,3-b]pyridazin-6-amine, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
5LYK
 
 | | CRYSTAL STRUCTURE OF INTRACELLULAR B30.2 DOMAIN OF BTN3A1 BOUND TO CITRATE | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 3 member A1, CITRATE ANION | | Authors: | Mohammed, F, Baker, A.T, Salim, M, Willcox, B.E. | | Deposit date: | 2016-09-28 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | BTN3A1 Discriminates gamma delta T Cell Phosphoantigens from Nonantigenic Small Molecules via a Conformational Sensor in Its B30.2 Domain.
ACS Chem. Biol., 12, 2017
|
|
5NQV
 
 | | Structure of the Arabidopsis Thaliana TOPLESS N-terminal domain | | Descriptor: | EAR motif of IAA27, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Nanao, M.H, Arevalillo, M.R, Vinos-Poyo, T, Parcy, F, Dumas, R. | | Deposit date: | 2017-04-21 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Arabidopsis TOPLESS corepressor provides insight into the evolution of transcriptional repression.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5NQS
 
 | | Structure of the Arabidopsis Thaliana TOPLESS N-terminal domain | | Descriptor: | Protein TOPLESS | | Authors: | Nanao, M.H, Arevalillo, M.R, Vinos-Poyo, T, Parcy, F, Dumas, R. | | Deposit date: | 2017-04-21 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of the Arabidopsis TOPLESS corepressor provides insight into the evolution of transcriptional repression.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4TW1
 
 | | Crystal structure of the octameric pore complex of the Staphylococcus aureus Bi-component Toxin LukGH | | Descriptor: | Possible leukocidin subunit | | Authors: | Logan, D.T, Hakansson, M, Saline, M, Kimbung, R, Badarau, A, Rouha, H, Nagy, E. | | Deposit date: | 2014-06-29 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Function Analysis of Heterodimer Formation, Oligomerization, and Receptor Binding of the Staphylococcus aureus Bi-component Toxin LukGH.
J.Biol.Chem., 290, 2015
|
|
3FRH
 
 | | Structure of the 16S rRNA methylase RmtB, P21 | | Descriptor: | 16S rRNA methylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schmitt, E, Galimand, M, Panvert, M, Dupechez, M, Courvalin, P, Mechulam, Y. | | Deposit date: | 2009-01-08 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural bases for 16 S rRNA methylation catalyzed by ArmA and RmtB methyltransferases
J.Mol.Biol., 388, 2009
|
|
3FRI
 
 | | Structure of the 16S rRNA methylase RmtB, I222 | | Descriptor: | 16S rRNA methylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schmitt, E, Galimand, M, Panvert, M, Dupechez, M, Courvalin, P, Mechulam, Y. | | Deposit date: | 2009-01-08 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural bases for 16 S rRNA methylation catalyzed by ArmA and RmtB methyltransferases
J.Mol.Biol., 388, 2009
|
|
4INL
 
 | | Crystal structure of SplD protease from Staphylococcus aureus at 2.1 A resolution | | Descriptor: | Serine protease SplD | | Authors: | Cichon, P, Zdzalik, M, Kalinska, M, Wysocka, M, Stec-Niemczyk, J, Stennicke, H.R, Jabaiah, A, Markiewicz, M, Wladyka, B, Daugherty, P.S, Lesner, A, Rolka, K, Dubin, A, Potempa, J, Dubin, G. | | Deposit date: | 2013-01-04 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and Structural Characterization of SplD Protease from Staphylococcus aureus.
Plos One, 8, 2013
|
|
4RZD
 
 | | Crystal Structure of a PreQ1 Riboswitch | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, PreQ1-III Riboswitch (Class 3) | | Authors: | Wedekind, J.E, Liberman, J.A, Salim, M. | | Deposit date: | 2014-12-20 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural analysis of a class III preQ1 riboswitch reveals an aptamer distant from a ribosome-binding site regulated by fast dynamics.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4TRP
 
 | | Crystal structure of monoclonal antibody against neuroblastoma associated antigen. | | Descriptor: | Heavy chain of monoclonal antibody against neuroblastoma associated antigen, Light chain of monoclonal antibody against neuroblastoma associated antigen | | Authors: | Horwacik, I, Golik, P, Grudnik, P, Zdzalik, M, Rokita, H, Dubin, G. | | Deposit date: | 2014-06-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis of GD2 Ganglioside and Mimetic Peptide Recognition by 14G2a Antibody.
Mol.Cell Proteomics, 14, 2015
|
|
6G5G
 
 | | Crystal structure of an engineered Botulinum Neurotoxin type B mutant E1191M/S1199Y in complex with human synaptotagmin 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Masuyer, G, Elliot, M, Favre-Guilmard, C, Liu, S.M, Maignel, J, Beard, M, Carre, D, Kalinichev, M, Lezmi, S, Mir, I, Nicoleau, C, Palan, S, Perier, C, Raban, E, Dong, M, Krupp, J, Stenmark, P. | | Deposit date: | 2018-03-29 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineered botulinum neurotoxin B with improved binding to human receptors has enhanced efficacy in preclinical models.
Sci Adv, 5, 2019
|
|
