4AEC
 
 | | Crystal Structure of the Arabidopsis thaliana O-Acetyl-Serine-(Thiol)- Lyase C | | Descriptor: | ACETATE ION, CYSTEINE SYNTHASE, MITOCHONDRIAL, ... | | Authors: | Feldman-Salit, A, Wirtz, M, Lenherr, E.D, Throm, C, Hothorn, M, Scheffzek, K, Hell, R, Wade, R.C. | | Deposit date: | 2012-01-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosterically Gated Enzyme Dynamics in the Cysteine Synthase Complex Regulate Cysteine Biosynthesis in Arabidopsis Thaliana.
Structure, 20, 2012
|
|
4M65
 
 | | In situ thermolysin crystallized on a MiTeGen micromesh with asparagine ligand | | Descriptor: | 1,2-ETHANEDIOL, ASPARAGINE, CALCIUM ION, ... | | Authors: | Yin, X, Scalia, A, Leroy, L, Cuttitta, C.M, Polizzo, G.M, Ericson, D.L, Roessler, C.G, Campos, O, Agarwal, R, Allaire, M, Orville, A.M, Jackimowicz, R, Ma, M.Y, Sweet, R.M, Soares, A.S. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hitting the target: fragment screening with acoustic in situ
co-crystallization of proteins plus fragment libraries on
pin-mounted data-collection micromeshes
Acta Crystallogr.,Sect.D, D70
|
|
1TKF
 
 | | Streptomyces griseus aminopeptidase complexed with D-tryptophan | | Descriptor: | Aminopeptidase, CALCIUM ION, D-TRYPTOPHAN, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-06-08 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Interactions of D Amino Acids with Streptomyces griseus Aminopeptidase
To be Published
|
|
7S5J
 
 | |
4PA1
 
 | |
1TKJ
 
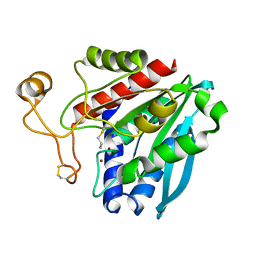 | | Streptomyces griseus aminopeptidase complexed with D-Methionine | | Descriptor: | Aminopeptidase, CALCIUM ION, D-METHIONINE, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-06-08 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Interactions of D Amino Acids with Streptomyces griseus Aminopeptidase
To be Published
|
|
7LP5
 
 | | Structure of Nedd4L WW3 domain | | Descriptor: | Angiomotin,E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Alam, S.L, Alian, A, Thompson, T, Rheinemann, L, Volkman, B.F, Peterson, F.C, Sundquist, W.I. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
1XBU
 
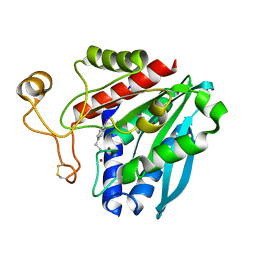 | | Streptomyces griseus aminopeptidase complexed with p-iodo-D-phenylalanine | | Descriptor: | Aminopeptidase, CALCIUM ION, P-IODO-D-PHENYLALANINE, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-08-31 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Streptomyces griseus aminopeptidase complexed with p-iodo-D-phenylalanine
To be Published
|
|
1QG0
 
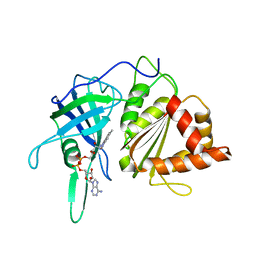 | | WILD-TYPE PEA FNR | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE) | | Authors: | Deng, Z, Aliverti, A, Zanetti, G, Arakaki, A.K, Ottado, J, Orellano, E.G, Calcaterra, N.B, Ceccarelli, E.A, Carrillo, N, Karplus, P.A. | | Deposit date: | 1999-04-18 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A productive NADP+ binding mode of ferredoxin-NADP+ reductase revealed by protein engineering and crystallographic studies.
Nat.Struct.Biol., 6, 1999
|
|
1QGA
 
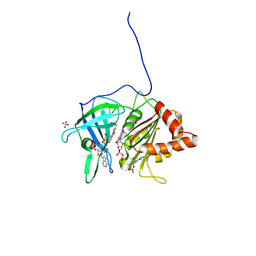 | | PEA FNR Y308W MUTANT IN COMPLEX WITH NADP+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE), ... | | Authors: | Deng, Z, Aliverti, A, Zanetti, G, Arakaki, A.K, Ottado, J, Orellano, E.G, Calcaterra, N.B, Ceccarelli, E.A, Carrillo, N, Karplus, P.A. | | Deposit date: | 1999-04-18 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A productive NADP+ binding mode of ferredoxin-NADP+ reductase revealed by protein engineering and crystallographic studies.
Nat.Struct.Biol., 6, 1999
|
|
7N87
 
 | |
7U5O
 
 | | CRYSTAL STRUCTURE OF THE BONE MORPHOGENETIC PROTEIN RECEPTOR TYPE 2 LIGAND BINDING DOMAIN IN COMPLEX WITH ACTIVIN-B | | Descriptor: | Bone morphogenetic protein receptor type-2, Inhibin beta B chain | | Authors: | Chu, K.Y, Malik, A, Thamilselvan, V, Martinez-Hackert, E. | | Deposit date: | 2022-03-02 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Type II BMP and activin receptors BMPR2 and ACVR2A share a conserved mode of growth factor recognition.
J.Biol.Chem., 298, 2022
|
|
7U5P
 
 | | CRYSTAL STRUCTURE OF THE ACTIVIN RECEPTOR TYPE-2A LIGAND BINDING DOMAIN IN COMPLEX WITH ACTIVIN-A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2A, Inhibin beta A chain | | Authors: | Chu, K.Y, Malik, A, Thamilselvan, V, Martinez-Hackert, E. | | Deposit date: | 2022-03-02 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Type II BMP and activin receptors BMPR2 and ACVR2A share a conserved mode of growth factor recognition.
J.Biol.Chem., 298, 2022
|
|
5EU7
 
 | | Crystal structure of HIV-1 integrase catalytic core in complex with Fab | | Descriptor: | FAB Heavy Chain, FAB light chain, Integrase | | Authors: | Galilee, M, Griner, S.L, Stroud, R.M, Alian, A. | | Deposit date: | 2015-11-18 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The Preserved HTH-Docking Cleft of HIV-1 Integrase Is Functionally Critical.
Structure, 24, 2016
|
|
5DCK
 
 | |
1TKH
 
 | | Streptomyces griseus aminopeptidase complexed with D-Phenylalanine | | Descriptor: | Aminopeptidase, CALCIUM ION, D-PHENYLALANINE, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-06-08 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Interactions of D Amino Acids with Streptomyces griseus Aminopeptidase
To be Published
|
|
2PMO
 
 | | Crystal structure of PfPK7 in complex with hymenialdisine | | Descriptor: | 4-(5-AMINO-4-OXO-4H-PYRAZOL-3-YL)-2-BROMO-4,5,6,7-TETRAHYDRO-3AH-PYRROLO[2,3-C]AZEPIN-8-ONE, Ser/Thr protein kinase | | Authors: | Merckx, A, Echalier, A, Noble, M, Endicott, J. | | Deposit date: | 2007-04-23 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of P. falciparum protein kinase 7 identify an activation motif and leads for inhibitor design.
Structure, 16, 2008
|
|
2Q2G
 
 | | Crystal structure of dimerization domain of HSP40 from Cryptosporidium parvum, cgd2_1800 | | Descriptor: | Heat shock 40 kDa protein, putative (fragment), SULFATE ION | | Authors: | Wernimont, A.K, Lew, J, Lin, L, Hassanali, A, Kozieradzki, I, Wasney, G, Vedadi, M, Walker, J.R, Zhao, Y, Schapira, M, Bochkarev, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Hui, R, Brokx, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-28 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dimerization domain of HSP40 from Cryptosporidium parvum, cgd2_1800.
To be Published
|
|
2PLU
 
 | | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_4120 | | Descriptor: | 20k cyclophilin, putative | | Authors: | Wernimont, A.K, Lew, J, Hills, T, Kozieradzki, I, Lin, Y.H, Hassanali, A, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_4120.
To be Published
|
|
2PMN
 
 | | Crystal structure of PfPK7 in complex with an ATP-site inhibitor | | Descriptor: | 4-(6-{[(1S)-1-(HYDROXYMETHYL)-2-METHYLPROPYL]AMINO}IMIDAZO[1,2-B]PYRIDAZIN-3-YL)BENZONITRILE, Ser/Thr protein kinase, putative | | Authors: | Merckx, A, Echalier, A, Noble, M, Endicott, J. | | Deposit date: | 2007-04-23 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of P. falciparum protein kinase 7 identify an activation motif and leads for inhibitor design.
Structure, 16, 2008
|
|
2Q0V
 
 | | Crystal structure of ubiquitin conjugating enzyme E2, putative, from Plasmodium falciparum | | Descriptor: | PHOSPHATE ION, Ubiquitin-conjugating enzyme E2, putative | | Authors: | Wernimont, A.K, Lew, J, Hassanali, A, Lin, L, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Brokx, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-22 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of ubiquitin conjugating enzyme E2, putative, from Plasmodium falciparum.
To be Published
|
|
7M2Y
 
 | | Closed conformation of the Yeast wild-type gamma-TuRC | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Spindle pole body component 110, Spindle pole body component SPC97, ... | | Authors: | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | Deposit date: | 2021-03-17 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
7M2X
 
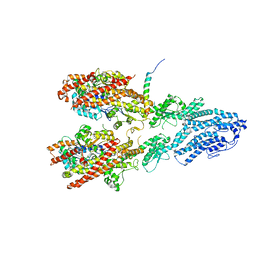 | | Open conformation of the Yeast wild-type gamma-TuRC | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Spindle pole body component 110, Spindle pole body component SPC97, ... | | Authors: | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | Deposit date: | 2021-03-17 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
7M2W
 
 | | Engineered disulfide cross-linked closed conformation of the Yeast gamma-TuRC(SS) | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Spindle pole body component 110, Spindle pole body component SPC97, ... | | Authors: | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | Deposit date: | 2021-03-17 | | Release date: | 2021-05-12 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
7M2Z
 
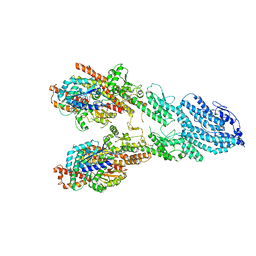 | | Monomeric single-particle reconstruction of the Yeast gamma-TuSC | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Spindle pole body component SPC97, Spindle pole body component SPC98, ... | | Authors: | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | Deposit date: | 2021-03-17 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
