3P6B
 
 | | The crystal structure of CelK CBM4 from Clostridium thermocellum | | Descriptor: | ACETATE ION, CALCIUM ION, Cellulose 1,4-beta-cellobiosidase, ... | | Authors: | Alahuhta, P.M, Luo, Y, Lunin, V.V. | | Deposit date: | 2010-10-11 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of CBM4 from Clostridium thermocellum cellulase K.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3NS7
 
 | |
3T9G
 
 | | The crystal structure of family 3 pectate lyase from Caldicellulosiruptor bescii | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2011-08-02 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A 1.5 A resolution X-ray structure of the catalytic module of Caldicellulosiruptor bescii family 3 pectate lyase.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3PDG
 
 | |
3PDD
 
 | |
4EL8
 
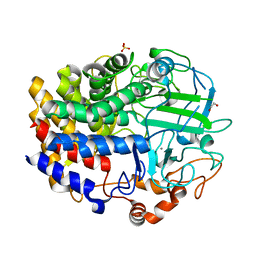 | | The unliganded structure of C.bescii CelA GH48 module | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glycoside hydrolase family 48, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2012-04-10 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Revealing nature's cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA.
Science, 342, 2013
|
|
1XD3
 
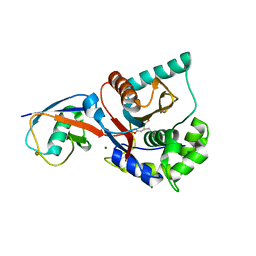 | | Crystal structure of UCHL3-UbVME complex | | Descriptor: | MAGNESIUM ION, METHYL 4-AMINOBUTANOATE, UBC protein, ... | | Authors: | Misaghi, S, Galardy, P.J, Meester, W.J.N, Ovaa, H, Ploegh, H.L, Gaudet, R. | | Deposit date: | 2004-09-03 | | Release date: | 2004-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the Ubiquitin Hydrolase UCH-L3 Complexed with a Suicide Substrate
J.Biol.Chem., 280, 2005
|
|
3VGN
 
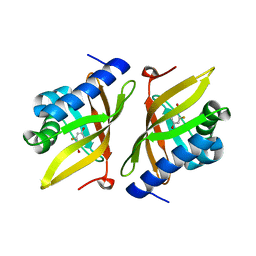 | | Crystal Structure of Ketosteroid Isomerase D40N from Pseudomonas putida (pKSI) with bound 3-fluoro-4-nitrophenol | | Descriptor: | 3-fluoro-4-nitrophenol, Steroid Delta-isomerase | | Authors: | Caaveiro, J.M.M, Pybus, B, Ringe, D, Petsko, G.A, Sigala, P.A. | | Deposit date: | 2011-08-16 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Quantitative dissection of hydrogen bond-mediated proton transfer in the ketosteroid isomerase active site
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4WD4
 
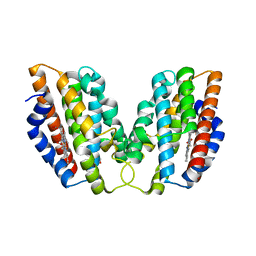 | | Crystal structure of human HO1 H25R | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Caaveiro, J.M.M, Morante, K, Sigala, P, Tsumoto, K. | | Deposit date: | 2014-09-06 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | In-Cell Enzymology To Probe His-Heme Ligation in Heme Oxygenase Catalysis
Biochemistry, 55, 2016
|
|
1XDS
 
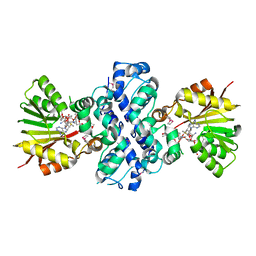 | | Crystal structure of Aclacinomycin-10-hydroxylase (RdmB) in complex with S-adenosyl-L-methionine (SAM) and 11-deoxy-beta-rhodomycin (DbrA) | | Descriptor: | 11-DEOXY-BETA-RHODOMYCIN, Protein RdmB, S-ADENOSYLMETHIONINE | | Authors: | Jansson, A, Koskiniemi, H, Erola, A, Wang, J, Mantsala, P, Schneider, G, Niemi, J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-09-08 | | Release date: | 2004-11-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Aclacinomycin 10-Hydroxylase Is a Novel Substrate-assisted Hydroxylase Requiring S-Adenosyl-L-methionine as Cofactor
J.Biol.Chem., 280, 2005
|
|
2VEU
 
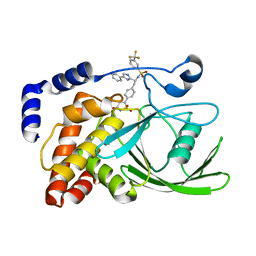 | | Crystal structure of protein tyrosine phosphatase 1B in complex with an isothiazolidinone-containing inhibitor | | Descriptor: | N-[(1S)-2-{4-[(5S)-1,1-dioxido-3-oxoisothiazolidin-5-yl]phenyl}-1-(4-phenyl-1H-imidazol-2-yl)ethyl]-3-(trifluoromethyl)benzenesulfonamide, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Douty, B, Wayland, B, Ala, P.J, Bower, M.J, Pruitt, J, Bostrom, L, Wei, M, Klabe, R, Gonneville, L, Wynn, R, Burn, T.C, Liu, P.C.C, Combs, A.P, Yue, E.W. | | Deposit date: | 2007-10-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Isothiazolidinone Inhibitors of Ptp1B Containing Imidazoles and Imidazolines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VEW
 
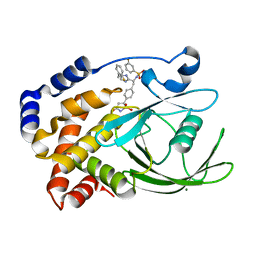 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B IN COMPLEX WITH AN ISOTHIAZOLIDINONE-CONTAINING INHIBITOR | | Descriptor: | 3-fluoro-N-[(1S)-1-[4-[(2-fluorophenyl)methyl]imidazol-2-yl]-2-[4-[(5S)-1,1,3-trioxo-1,2-thiazolidin-5-yl]phenyl]ethyl]benzenesulfonamide, MAGNESIUM ION, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Douty, B, Wayland, B, Ala, P.J, Bower, M.J, Pruitt, J, Bostrom, L, Wei, M, Klabe, R, Gonneville, L, Wynn, R, Burn, T.C, Liu, P.C.C, Combs, A.P, Yue, E.W. | | Deposit date: | 2007-10-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Isothiazolidinone Inhibitors of Ptp1B Containing Imidazoles and Imidazolines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VEX
 
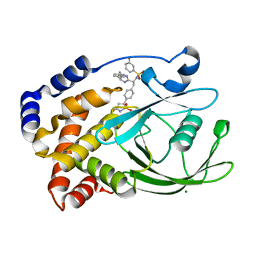 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B IN COMPLEX WITH AN ISOTHIAZOLIDINONE-CONTAINING INHIBITOR | | Descriptor: | MAGNESIUM ION, N-{(1S)-2-{4-[(5S)-1,1-dioxido-3-oxoisothiazolidin-5-yl]phenyl}-1-[(4R)-4-(2-phenylethyl)-4,5-dihydro-1H-imidazol-2-yl]ethyl}-3-fluorobenzenesulfonamide, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Douty, B, Wayland, B, Ala, P.J, Bower, M.J, Pruitt, J, Bostrom, L, Wei, M, Klabe, R, Gonneville, L, Wynn, R, Burn, T.C, Liu, P.C.C, Combs, A.P, Yue, E.W. | | Deposit date: | 2007-10-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isothiazolidinone Inhibitors of Ptp1B Containing Imidazoles and Imidazolines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VEV
 
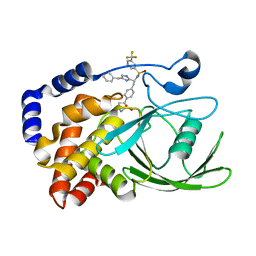 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B IN COMPLEX WITH AN ISOTHIAZOLIDINONE-CONTAINING INHIBITOR | | Descriptor: | MAGNESIUM ION, N-[(1S)-1-(4-benzyl-1H-imidazol-2-yl)-2-{4-[(5S)-1,1-dioxido-3-oxoisothiazolidin-5-yl]phenyl}ethyl]-3-(trifluoromethyl)benzenesulfonamide, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Douty, B, Wayland, B, Ala, P.J, Bower, M.J, Pruitt, J, Bostrom, L, Wei, M, Klabe, R, Gonneville, L, Wynn, R, Burn, T.C, Liu, P.C.C, Combs, A.P, Yue, E.W. | | Deposit date: | 2007-10-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isothiazolidinone Inhibitors of Ptp1B Containing Imidazoles and Imidazolines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VEY
 
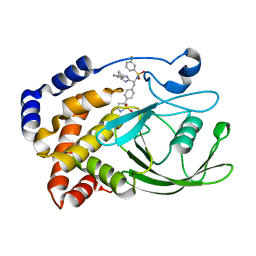 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B IN COMPLEX WITH AN ISOTHIAZOLIDINONE-CONTAINING INHIBITOR | | Descriptor: | N-{(1S)-2-{4-[(5S)-1,1-dioxido-3-oxoisothiazolidin-5-yl]phenyl}-1-[4-(3-phenylpropyl)-1H-imidazol-2-yl]ethyl}-3-fluorobenzenesulfonamide, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Douty, B, Wayland, B, Ala, P.J, Bower, M.J, Pruitt, J, Bostrom, L, Wei, M, Klabe, R, Gonneville, L, Wynn, R, Burn, T.C, Liu, P.C.C, Combs, A.P, Yue, E.W. | | Deposit date: | 2007-10-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isothiazolidinone Inhibitors of Ptp1B Containing Imidazoles and Imidazolines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1XDU
 
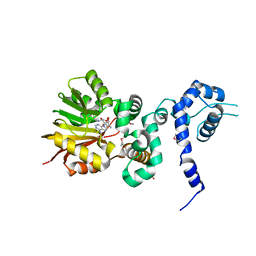 | | Crystal structure of Aclacinomycin-10-hydroxylase (RdmB) in complex with Sinefungin (SFG) | | Descriptor: | ACETATE ION, Protein RdmB, SINEFUNGIN | | Authors: | Jansson, A, Koskiniemi, H, Erola, A, Wang, J, Mantsala, P, Schneider, G, Niemi, J. | | Deposit date: | 2004-09-08 | | Release date: | 2004-11-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aclacinomycin 10-Hydroxylase Is a Novel Substrate-assisted Hydroxylase Requiring S-Adenosyl-L-methionine as Cofactor
J.Biol.Chem., 280, 2005
|
|
7Q8B
 
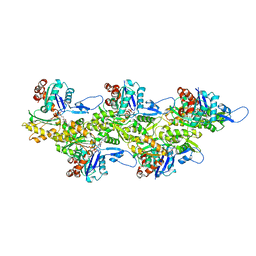 | | Leishmania major actin filament in ADP-Pi state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, MAGNESIUM ION, ... | | Authors: | Kotila, T, Muniyandi, S, Lappalainen, P, Huiskonen, J.T. | | Deposit date: | 2021-11-11 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of rapid actin dynamics in the evolutionarily divergent Leishmania parasite.
Nat Commun, 13, 2022
|
|
7NW1
 
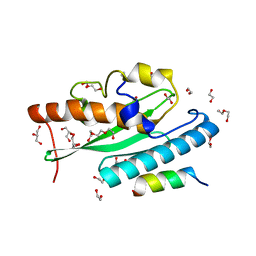 | | Crystal structure of UFC1 in complex with UBA5 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Manoj Kumar, P, Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2021-03-16 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for UFM1 transfer from UBA5 to UFC1.
Nat Commun, 12, 2021
|
|
7Q8C
 
 | | Leishmania major actin filament in ADP-state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, MAGNESIUM ION | | Authors: | Kotila, T, Muniyandi, S, Lappalainen, P, Huiskonen, J.T. | | Deposit date: | 2021-11-11 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural basis of rapid actin dynamics in the evolutionarily divergent Leishmania parasite.
Nat Commun, 13, 2022
|
|
7Q8S
 
 | | Leishmania major ADP-actin filament decorated with Leishmania major cofilin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADF/Cofilin, Actin, ... | | Authors: | Kotila, T, Muniyandi, S, Lappalainen, P, Huiskonen, J.T. | | Deposit date: | 2021-11-11 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of rapid actin dynamics in the evolutionarily divergent Leishmania parasite.
Nat Commun, 13, 2022
|
|
7NVJ
 
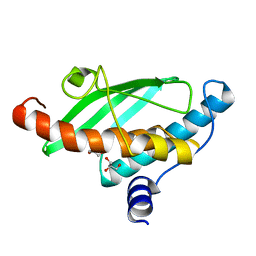 | | Crystal structure of UFC1 Y110A & F121A | | Descriptor: | GLYCEROL, Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Manoj Kumar, P, Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2021-03-15 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for UFM1 transfer from UBA5 to UFC1.
Nat Commun, 12, 2021
|
|
7NVK
 
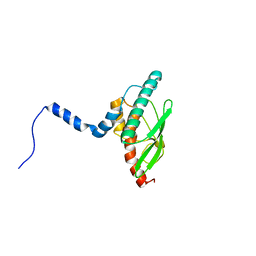 | | Crystal structure of UBA5 fragment fused to the N-terminus of UFC1 | | Descriptor: | Ubiquitin-like modifier-activating enzyme 5,Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Manoj Kumar, P, Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2021-03-15 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structural basis for UFM1 transfer from UBA5 to UFC1.
Nat Commun, 12, 2021
|
|
7ZTD
 
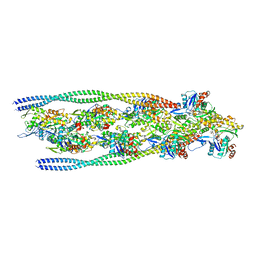 | | Non-muscle F-actin decorated with non-muscle tropomyosin 3.2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Non-muscle tropomyosin 3.2, actin, ... | | Authors: | Selvaraj, M, Kokate, S, Kogan, K, Kotila, T, Kremneva, E, Lappalainen, P, Huiskonen, J.T. | | Deposit date: | 2022-05-09 | | Release date: | 2023-01-11 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis underlying specific biochemical activities of non-muscle tropomyosin isoforms.
Cell Rep, 42, 2023
|
|
8AIJ
 
 | | STRUCTURE OF THE LECB LECTIN FROM PSEUDOMONAS AERUGINOSA STRAIN PAO1 IN COMPLEX WITH N-(alpha-L-Fucopyranosyl)benzamide (6) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Fucose-binding lectin PA-IIL, ... | | Authors: | Meiers, J, Mala, P, Varrot, A, Siebs, E, Imberty, A, Titz, A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of N -beta-l-Fucosyl Amides as High-Affinity Ligands for the Pseudomonas aeruginosa Lectin LecB.
J.Med.Chem., 65, 2022
|
|
8AIY
 
 | | STRUCTURE OF THE LECB LECTIN FROM PSEUDOMONAS AERUGINOSA STRAIN PAO1 IN COMPLEX WITH N-(beta-L-Fucopyranosyl)-biphenyl-3-carboxamide (4i) | | Descriptor: | CALCIUM ION, Fucose-binding lectin PA-IIL, N-(beta-L-Fucopyranosyl)-biphenyl-3-carboxamide, ... | | Authors: | Meiers, J, Mala, P, Varrot, A, Siebs, E, Imberty, A, Titz, A. | | Deposit date: | 2022-07-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of N -beta-l-Fucosyl Amides as High-Affinity Ligands for the Pseudomonas aeruginosa Lectin LecB.
J.Med.Chem., 65, 2022
|
|
