2LMK
 
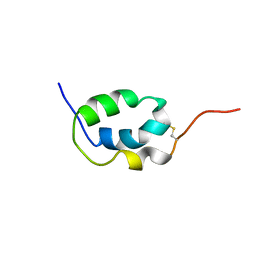 | | Solution Structure of Mouse Pheromone ESP1 | | Descriptor: | Exocrine gland-secreting peptide 1 | | Authors: | Yoshinaga, S, Sato, T, Hirakane, M, Esaki, K, Hamaguchi, T, Haga-Yamanaka, S, Tsunoda, M, Kimoto, H, Shimada, I, Touhara, K, Terasawa, H. | | Deposit date: | 2011-12-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the Mouse Sex Peptide Pheromone ESP1 Reveals a Molecular Basis for Specific Binding to the Class-C G-Protein-Coupled Vomeronasal Receptor
J.Biol.Chem., 2013
|
|
4Y9H
 
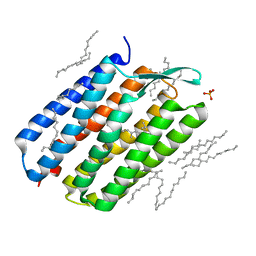 | | The 1.43 angstrom crystal structure of bacteriorhodopsin crystallized from bicelles | | Descriptor: | Bacteriorhodopsin, DECANE, DODECANE, ... | | Authors: | Saiki, H, Sugiyama, S, Kakinouchi, K, Kawatake, S, Hanashima, S, Matsumori, N, Murata, M. | | Deposit date: | 2015-02-17 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The 1.43 angstrom crystal structure of bacteriorhodopsin crystallized from bicelles
To Be Published
|
|
5TT4
 
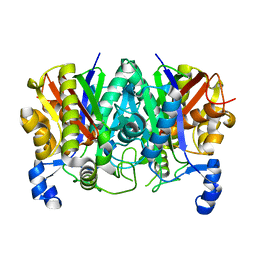 | | Determining the Molecular Basis For Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase | | Authors: | Jackson, D.R, Valentic, T.R, Patel, A, Tsai, S.C, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2016-11-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determining the Molecular Basis For Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
3W3E
 
 | |
3W3D
 
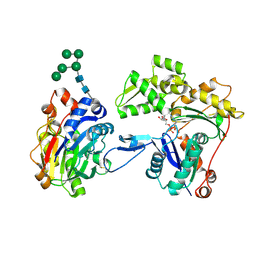 | | Crystal structure of smooth muscle G actin DNase I complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, gamma-enteric smooth muscle, ... | | Authors: | Sakabe, N, Sakabe, K, Sasaki, K, Kondo, H, Shimomur, M. | | Deposit date: | 2012-12-20 | | Release date: | 2013-01-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined structure and solvent network of chicken gizzard G-actin DNase 1 complex at 1.8A resolution
Acta Crystallogr.,Sect.A, 49, 1993
|
|
3W7Z
 
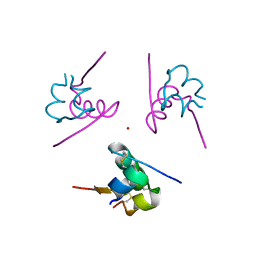 | |
2RUJ
 
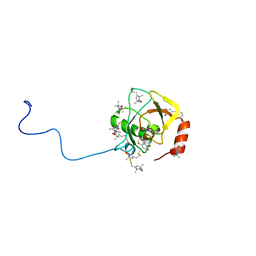 | | Solution structure of MTSL spin-labeled Schizosaccharomyces pombe Sin1 CRIM domain | | Descriptor: | Stress-activated map kinase-interacting protein 1 | | Authors: | Furuita, K, Kataoka, S, Sugiki, T, Kobayashi, N, Ikegami, T, Shiozaki, K, Fujiwara, T, Kojima, C. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-29 | | Method: | SOLUTION NMR | | Cite: | Utilization of paramagnetic relaxation enhancements for high-resolution NMR structure determination of a soluble loop-rich protein with sparse NOE distance restraints
J.Biomol.Nmr, 61, 2015
|
|
2RVK
 
 | |
7WU9
 
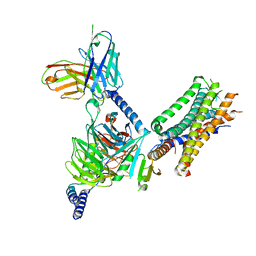 | | Cryo-EM structure of the human EP3-Gi signaling complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Suno, R, Sugita, Y, Morimoto, K, Iwasaki, K, Kato, T, Kobayashi, T. | | Deposit date: | 2022-02-07 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.375 Å) | | Cite: | Structural insights into the G protein selectivity revealed by the human EP3-G i signaling complex.
Cell Rep, 40, 2022
|
|
3EFD
 
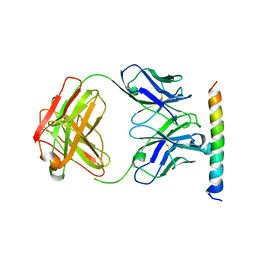 | | The crystal structure of the cytoplasmic domain of KcsA | | Descriptor: | FabH, FabL, KcsA | | Authors: | Uysal, S, Vasquez, V, Tereshko, V, Esaki, K, Fellouse, F.A, Sidhu, S.S, Koide, S, Perozo, E, Kossiakoff, A. | | Deposit date: | 2008-09-08 | | Release date: | 2009-04-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of full-length KcsA in its closed conformation.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3W7Y
 
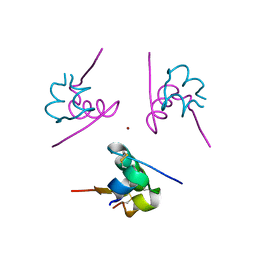 | |
3WMG
 
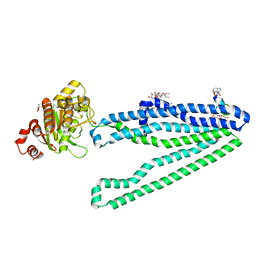 | | Crystal structure of an inward-facing eukaryotic ABC multidrug transporter G277V/A278V/A279V mutant in complex with an cyclic peptide inhibitor, aCAP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP-binding cassette, sub-family B, ... | | Authors: | Kodan, A, Yamaguchi, T, Nakatsu, T, Sakiyama, K, Hipolito, C.J, Fujioka, A, Hirokane, R, Ikeguchi, K, Watanabe, B, Hirtake, J, Kimura, Y, Suga, H, Ueda, K, Kato, H. | | Deposit date: | 2013-11-18 | | Release date: | 2014-04-30 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WXQ
 
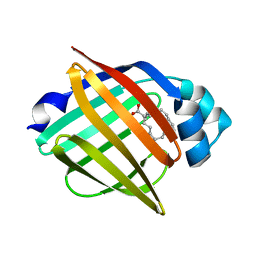 | | Serial femtosecond X-ray structure of human fatty acid-binding protein type-3 (FABP3) in complex with stearic acid (C18:0) determined using X-ray free-electron laser at SACLA | | Descriptor: | Fatty acid-binding protein, heart, STEARIC ACID | | Authors: | Mizohata, E, Suzuki, M, Kakinouchi, K, Sugiyama, S, Murata, M, Sugahara, M, Nango, E, Tanaka, T, Tanaka, R, Tono, K, Song, C, Hatsui, T, Joti, Y, Yabashi, M, Iwata, S. | | Deposit date: | 2014-08-04 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat. Methods, 12, 2015
|
|
3X0V
 
 | | Structure of L-lysine oxidase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, L-lysine oxidase | | Authors: | Sano, T, Uchida, Y, Amano, M, Kawaguchi, T, Kondo, H, Inagaki, K, Imada, K. | | Deposit date: | 2014-10-22 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recombinant expression, molecular characterization and crystal structure of antitumor enzyme, l-lysine alpha-oxidase from Trichoderma viride.
J.Biochem., 157, 2015
|
|
3UG3
 
 | | Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, ... | | Authors: | Im, D.-H, Miyazaki, K, Wakagi, T, Fushinobu, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Glycoside Hydrolase Family 51 alpha-L-Arabinofuranosidase from Thermotoga maritima
Biosci.Biotechnol.Biochem., 76, 2012
|
|
1OD5
 
 | | Crystal structure of glycinin A3B4 subunit homohexamer | | Descriptor: | CARBONATE ION, GLYCININ, MAGNESIUM ION | | Authors: | Adachi, M, Kanamori, J, Masuda, T, Yagasaki, K, Kitamura, K, Mikami, B, Utsumi, S. | | Deposit date: | 2003-02-13 | | Release date: | 2003-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Soybean 11S Globulin: Glycinin A3B4 Homohexamer
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1UD3
 
 | | Crystal structure of AmyK38 N289H mutant | | Descriptor: | SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UD8
 
 | | Crystal structure of AmyK38 with lithium ion | | Descriptor: | SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
3ADM
 
 | | Crystal structure of (Pro-Pro-Gly)4-Hyp-Ser-Gly-(Pro-Pro-Gly)4 | | Descriptor: | collagen-like peptide | | Authors: | Okuyama, K, Miyama, K, Masakiyo, K, Mizuno, K, Bachinger, H.P. | | Deposit date: | 2010-01-22 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Stabilization of triple-helical structures of collagen peptides containing a Hyp-Thr-Gly, Hyp-Val-Gly, or Hyp-Ser-Gly sequence.
Biopolymers, 95, 2011
|
|
1UD5
 
 | | Crystal structure of AmyK38 with rubidium ion | | Descriptor: | RUBIDIUM ION, SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UD4
 
 | | Crystal structure of calcium free alpha amylase from Bacillus sp. strain KSM-K38 (AmyK38, in calcium containing solution) | | Descriptor: | SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UD2
 
 | | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) | | Descriptor: | GLYCEROL, SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1G01
 
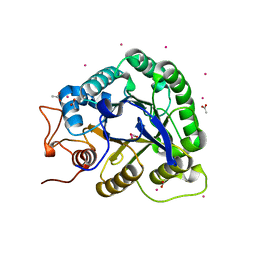 | | ALKALINE CELLULASE K CATALYTIC DOMAIN | | Descriptor: | ACETIC ACID, CADMIUM ION, ENDOGLUCANASE | | Authors: | Shirai, T, Ishida, H, Noda, J, Yamane, T, Ozaki, K, Hakamada, Y, Ito, S. | | Deposit date: | 2000-10-05 | | Release date: | 2001-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of alkaline cellulase K: insight into the alkaline adaptation of an industrial enzyme.
J.Mol.Biol., 310, 2001
|
|
3UG4
 
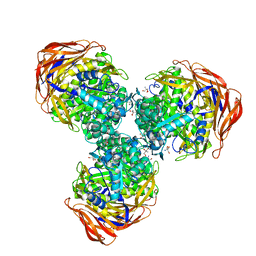 | | Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, alpha-L-arabinofuranose | | Authors: | Im, D.-H, Miyazaki, K, Wakagi, T, Fushinobu, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of Glycoside Hydrolase Family 51 alpha-L-Arabinofuranosidase from Thermotoga maritima
Biosci.Biotechnol.Biochem., 76, 2012
|
|
3UG5
 
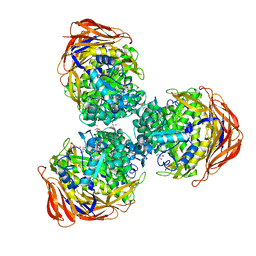 | | Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, beta-D-xylopyranose | | Authors: | Im, D.-H, Miyazaki, K, Wakagi, T, Fushinobu, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Glycoside Hydrolase Family 51 alpha-L-Arabinofuranosidase from Thermotoga maritima
Biosci.Biotechnol.Biochem., 76, 2012
|
|
