7DN2
 
 | | Acidic stable capsid structure of Helicobacter pylori bacteriophage KHP30 | | 分子名称: | Cement protein gp15, Major structural protein ORF14 | | 著者 | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | 登録日 | 2020-12-08 | | 公開日 | 2021-10-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Acid-stable capsid structure of Helicobacter pylori bacteriophage KHP30 by single-particle cryoelectron microscopy.
Structure, 30, 2022
|
|
7DOU
 
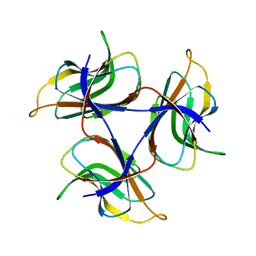 | | Trimeric cement protein structure of Helicobacter pylori bacteriophage KHP40 | | 分子名称: | Cement protein gp16 | | 著者 | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | 登録日 | 2020-12-17 | | 公開日 | 2021-10-27 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Acid-stable capsid structure of Helicobacter pylori bacteriophage KHP30 by single-particle cryoelectron microscopy.
Structure, 30, 2022
|
|
7DOD
 
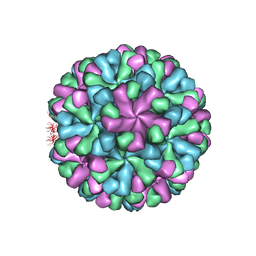 | | Capsid structure of human sapovirus | | 分子名称: | Calicivirin | | 著者 | Miyazaki, N, Murakami, K, Oka, T, Iwasaki, K, Katayama, K, Murata, K. | | 登録日 | 2020-12-14 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Atomic structure of human sapovirus capsid by single particle cryo-electron microscopy
To Be Published
|
|
3VKF
 
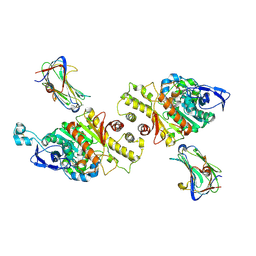 | | Crystal Structure of Neurexin 1beta/Neuroligin 1 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurexin-1-beta, ... | | 著者 | Tanaka, H, Miyazaki, N, Nogi, T, Iwasaki, K, Takagi, J. | | 登録日 | 2011-11-15 | | 公開日 | 2012-08-01 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Higher-order architecture of cell adhesion mediated by polymorphic synaptic adhesion molecules neurexin and neuroligin.
Cell Rep, 2, 2012
|
|
4P7W
 
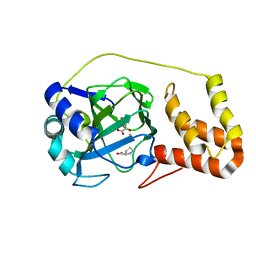 | | L-proline-bound L-proline cis-4-hydroxylase | | 分子名称: | 2-OXOGLUTARIC ACID, COBALT (II) ION, L-proline cis-4-hydroxylase, ... | | 著者 | Shomura, Y, Koketsu, K, Moriwaki, K, Hayashi, M, Mitsuhashi, S, Hara, R, Kino, K, Higuchi, Y. | | 登録日 | 2014-03-28 | | 公開日 | 2014-09-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Refined Regio- and Stereoselective Hydroxylation of l-Pipecolic Acid by Protein Engineering of l-Proline cis-4-Hydroxylase Based on the X-ray Crystal Structure.
Acs Synth Biol, 4, 2015
|
|
3MPW
 
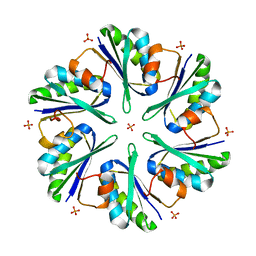 | | Structure of EUTM in 2-D protein membrane | | 分子名称: | Ethanolamine utilization protein eutM, PHOSPHATE ION | | 著者 | Sagermann, M, Takenoya, M, Nikolakakis, K. | | 登録日 | 2010-04-27 | | 公開日 | 2011-05-11 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystallographic insights into the pore structures and mechanisms of the EutL and EutM shell proteins of the ethanolamine-utilizing microcompartment of Escherichia coli.
J.Bacteriol., 192, 2010
|
|
7D6J
 
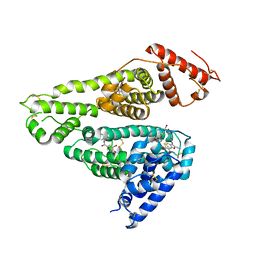 | | Human serum albumin complexed with benzbromarone | | 分子名称: | Serum albumin, [3,5-bis(bromanyl)-4-oxidanyl-phenyl]-(2-ethyl-1-benzofuran-3-yl)methanone | | 著者 | Kawai, A, Yamasaki, K. | | 登録日 | 2020-09-30 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.29 Å) | | 主引用文献 | Interaction of Benzbromarone with Subdomains IIIA and IB/IIA on Human Serum Albumin as the Primary and Secondary Binding Regions.
Mol Pharm., 18, 2021
|
|
2RUJ
 
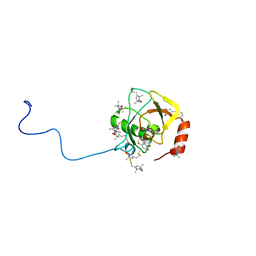 | | Solution structure of MTSL spin-labeled Schizosaccharomyces pombe Sin1 CRIM domain | | 分子名称: | Stress-activated map kinase-interacting protein 1 | | 著者 | Furuita, K, Kataoka, S, Sugiki, T, Kobayashi, N, Ikegami, T, Shiozaki, K, Fujiwara, T, Kojima, C. | | 登録日 | 2014-07-24 | | 公開日 | 2015-07-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Utilization of paramagnetic relaxation enhancements for high-resolution NMR structure determination of a soluble loop-rich protein with sparse NOE distance restraints
J.Biomol.Nmr, 61, 2015
|
|
6LFF
 
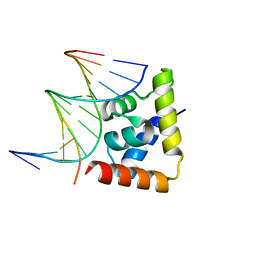 | | transcription factor SATB1 CUTr1 domain in complex with a phosphorothioate DNA | | 分子名称: | DNA (5'-D(*GP*(C7R)P*(PST)P*AP*AP*TP*AP*TP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*(AS)P*(PST)P*(AS)P*(PST)P*TP*AP*GP*C)-3'), DNA-binding protein SATB1 | | 著者 | Akutsu, Y, Kubota, T, Yamasaki, T, Yamasaki, K. | | 登録日 | 2019-12-02 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Enhanced affinity of racemic phosphorothioate DNA with transcription factor SATB1 arising from diastereomer-specific hydrogen bonds and hydrophobic contacts.
Nucleic Acids Res., 48, 2020
|
|
2GCC
 
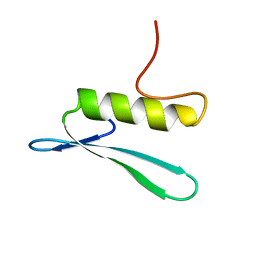 | | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, MINIMIZED MEAN STRUCTURE | | 分子名称: | ATERF1 | | 著者 | Allen, M.D, Yamasaki, K, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | 登録日 | 1998-03-13 | | 公開日 | 1999-03-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
3VP9
 
 | | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p mutant | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, General transcriptional corepressor TUP1 | | 著者 | Matsumura, H, Kusaka, N, Nakamura, T, Tanaka, N, Sagegami, K, Uegaki, K, Inoue, T, Mukai, Y. | | 登録日 | 2012-02-28 | | 公開日 | 2012-06-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.799 Å) | | 主引用文献 | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p and its functional implications
J.Biol.Chem., 287, 2012
|
|
7D4C
 
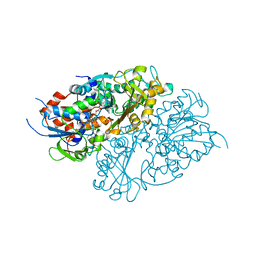 | | Structure of L-lysine oxidase precursor | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, L-Lysine alpha-oxidase, PHOSPHATE ION | | 著者 | Ito, N, Kitagawa, M, Matsumoto, Y, Inagaki, K, Imada, K. | | 登録日 | 2020-09-23 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural basis of enzyme activity regulation by the propeptide of l-lysine alpha-oxidase precursor from Trichoderma viride .
J Struct Biol X, 5, 2021
|
|
7D4D
 
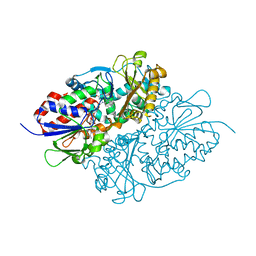 | | Structure of L-lysine oxidase precursor in complex with L-lysine (1.24M) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, L-Lysine alpha-oxidase, LYSINE | | 著者 | Kitagawa, M, Ito, N, Matsumoto, Y, Inagaki, K, Imada, K. | | 登録日 | 2020-09-23 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structural basis of enzyme activity regulation by the propeptide of l-lysine alpha-oxidase precursor from Trichoderma viride .
J Struct Biol X, 5, 2021
|
|
7D4E
 
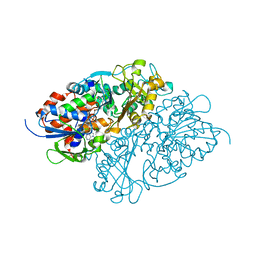 | | Structure of L-lysine oxidase precursor in complex with L-lysine (1.0 M) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, L-Lysine alpha-oxidase, LYSINE | | 著者 | Kitagawa, M, Ito, N, Matsumoto, Y, Inagaki, K, Imada, K. | | 登録日 | 2020-09-23 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural basis of enzyme activity regulation by the propeptide of l-lysine alpha-oxidase precursor from Trichoderma viride .
J Struct Biol X, 5, 2021
|
|
8GS6
 
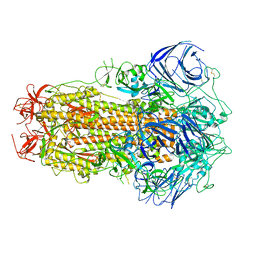 | | Structure of the SARS-CoV-2 BA.2.75 spike glycoprotein (closed state 1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Anraku, Y, Tabata-Sasaki, K, Kita, S, Fukuhara, H, Maenaka, K, Hashiguchi, T. | | 登録日 | 2022-09-05 | | 公開日 | 2022-10-26 | | 最終更新日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 Omicron BA.2.75 variant.
Cell Host Microbe, 30, 2022
|
|
3VK3
 
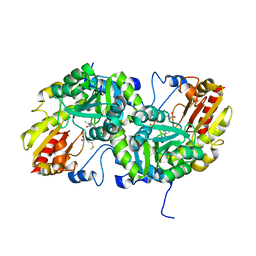 | | Crystal Structure of L-Methionine gamma-Lyase from Pseudomonas putida C116H Mutant Complexed with L-methionine | | 分子名称: | METHIONINE, Methionine gamma-lyase | | 著者 | Fukumoto, M, Kudou, D, Murano, S, Shiba, T, Sato, D, Tamura, T, Harada, S, Inagaki, K. | | 登録日 | 2011-11-07 | | 公開日 | 2012-09-19 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The role of amino acid residues in the active site of L-methionine gamma-lyase from Pseudomonas putida.
Biosci.Biotechnol.Biochem., 76, 2012
|
|
3VOV
 
 | | Crystal Structure of ROK Hexokinase from Thermus thermophilus | | 分子名称: | GLYCEROL, Glucokinase, ZINC ION | | 著者 | Nakamura, T, Kashima, Y, Mine, S, Oku, T, Uegaki, K. | | 登録日 | 2012-02-21 | | 公開日 | 2012-06-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Characterization and crystal structure of the thermophilic ROK hexokinase from Thermus thermophilus
J.Biosci.Bioeng., 2012
|
|
3VK4
 
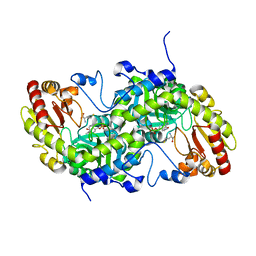 | | Crystal Structure of L-Methionine gamma-Lyase from Pseudomonas putida C116H Mutant complexed with L-homocysteine | | 分子名称: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Methionine gamma-lyase | | 著者 | Fukumoto, M, Kudou, D, Murano, S, Shiba, T, Sato, D, Tamura, T, Harada, S, Inagaki, K. | | 登録日 | 2011-11-07 | | 公開日 | 2012-09-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | The role of amino acid residues in the active site of L-methionine gamma-lyase from Pseudomonas putida.
Biosci.Biotechnol.Biochem., 76, 2012
|
|
6LXV
 
 | | Cryo-EM structure of phosphoketolase from Bifidobacterium longum | | 分子名称: | CALCIUM ION, Phosphoketolase, THIAMINE DIPHOSPHATE | | 著者 | Nakata, K, Miyazaki, N, Yamaguchi, H, Hirose, M, Miyano, H, Mizukoshi, T, Kashiwagi, T, Iwasaki, K. | | 登録日 | 2020-02-12 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.1 Å) | | 主引用文献 | High-resolution structure of phosphoketolase from Bifidobacterium longum determined by cryo-EM single-particle analysis.
J.Struct.Biol., 214, 2022
|
|
2RT4
 
 | |
6AB6
 
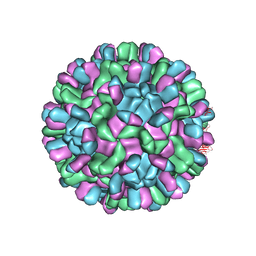 | | Cryo-EM structure of T=3 Penaeus vannamei nodavirus | | 分子名称: | CALCIUM ION, Capsid protein | | 著者 | Chen, N.C, Miyazaki, N, Yoshimura, M, Guan, H.H, Lin, C.C, Iwasaki, K, Chen, C.J. | | 登録日 | 2018-07-20 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
4D7Y
 
 | | Crystal structure of mouse C1QL1 globular domain | | 分子名称: | C1Q-RELATED FACTOR, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Kakegawa, W, Mitakidis, N, Miura, E, Abe, M, Matsuda, K, Takeo, Y, Kohda, K, Motohashi, J, Takahashi, A, Nagao, S, Muramatsu, S, Watanabe, M, Sakimura, K, Aricescu, A.R, Yuzaki, M. | | 登録日 | 2014-12-01 | | 公開日 | 2015-01-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Anterograde C1Ql1 Signaling is Required in Order to Determine and Maintain a Single-Winner Climbing Fiber in the Mouse Cerebellum
Neuron, 85, 2015
|
|
7X30
 
 | | Capsid structure of Staphylococcus jumbo bacteriophage S6 | | 分子名称: | Hoc-like protein ORF90, Major structural protein ORF12 | | 著者 | Koibuchi, W, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | 登録日 | 2022-02-27 | | 公開日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Capsid structure of Staphylococcus jumbo bacteriophage S6
To Be Published
|
|
7WMP
 
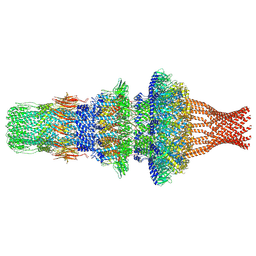 | | Tail structure of Helicobacter pylori bacteriophage KHP30 | | 分子名称: | Adaptor protein gp12, Nozzle protein gp25, Portal protein | | 著者 | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | 登録日 | 2022-01-15 | | 公開日 | 2023-03-01 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structure of Helicobacter pylori bacteriophage KHP30
To Be Published
|
|
5AWW
 
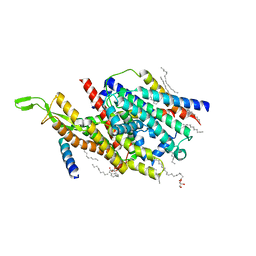 | | Precise Resting State of Thermus thermophilus SecYEG | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Protein translocase subunit SecE, Protein translocase subunit SecY, ... | | 著者 | Tanaka, Y, Sugano, Y, Takemoto, M, Kusakizako, T, Kumazaki, K, Ishitani, R, Nureki, O, Tsukazaki, T. | | 登録日 | 2015-07-10 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.724 Å) | | 主引用文献 | Crystal Structures of SecYEG in Lipidic Cubic Phase Elucidate a Precise Resting and a Peptide-Bound State.
Cell Rep, 13, 2015
|
|
