6UMW
 
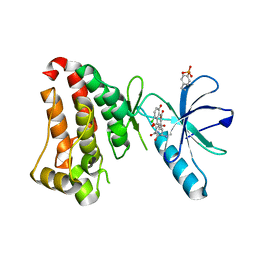 | | Crystal structure of hEphB1 bound with chlortetracycline | | Descriptor: | 7-CHLOROTETRACYCLINE, Ephrin type-B receptor 1 | | Authors: | Ahmed, M, Wang, P, Sadek, H. | | Deposit date: | 2019-10-10 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Identification of tetracycline combinations as EphB1 tyrosine kinase inhibitors for treatment of neuropathic pain.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KPL
 
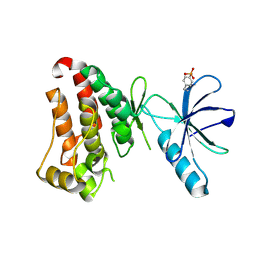 | | Crystal structure of hEphB1 in apo form | | Descriptor: | Ephrin type-B receptor 1 | | Authors: | Ahmed, M, Wang, P, Sadek, H. | | Deposit date: | 2020-11-11 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Identification of tetracycline combinations as EphB1 tyrosine kinase inhibitors for treatment of neuropathic pain.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KPM
 
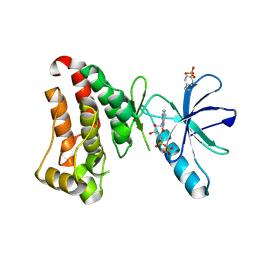 | | Crystal structure of hEphB1 bound with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ephrin type-B receptor 1 | | Authors: | Ahmed, M, Wang, P, Sadek, H. | | Deposit date: | 2020-11-11 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.608 Å) | | Cite: | Identification of tetracycline combinations as EphB1 tyrosine kinase inhibitors for treatment of neuropathic pain.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5CMA
 
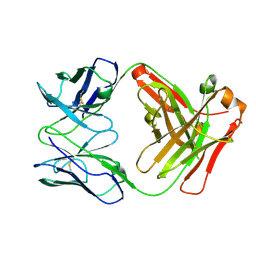 | | Anti-B7-H3 monoclonal antibody ch8H9 Fab fragment | | Descriptor: | Antibody ch8H9 Fab heavy chain, Antibody ch8H9 Fab light chain | | Authors: | Ahmed, M, Goldgur, Y, Cheng, M, Cheung, N.K. | | Deposit date: | 2015-07-16 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Humanized Affinity-matured Monoclonal Antibody 8H9 Has Potent Antitumor Activity and Binds to FG Loop of Tumor Antigen B7-H3.
J.Biol.Chem., 290, 2015
|
|
3VFG
 
 | |
4MDC
 
 | | Crystal structure of glutathione S-transferase from Sinorhizobium meliloti 1021, NYSGRC target 021389 | | Descriptor: | GLYCEROL, Putative glutathione S-transferase | | Authors: | Shabalin, I.G, Bacal, P, Cooper, D.R, Stead, M, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-22 | | Release date: | 2013-09-04 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of glutathione S-transferase from Sinorhizobium meliloti 1021
To be Published
|
|
4WCZ
 
 | | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Tkaczuk, K.L, Cooper, D.R, Chapman, H.C, Niedzialkowska, E, Cymborowski, M.T, Hillerich, B.S, Stead, M, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-05 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans
to be published
|
|
8VTT
 
 | | Meis1 homeobox domain bound to neomycin fragment | | Descriptor: | Homeobox protein Meis1, RIBOSTAMYCIN, SULFATE ION | | Authors: | Tomchick, D.R, Ahmed, M.S, Nguyen, N.U.N, Sadek, H.A. | | Deposit date: | 2024-01-27 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification of FDA-approved drugs that induce heart regeneration in mammals.
Nat Cardiovasc Res, 3, 2024
|
|
8VTS
 
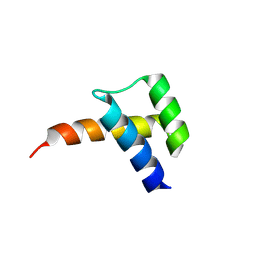 | | Meis1 homeobox domain bound to paromomycin fragment | | Descriptor: | 1,2-ETHANEDIOL, Homeobox protein Meis1, ISOPROPYL ALCOHOL, ... | | Authors: | Tomchick, D.R, Ahmed, M.S, Nguyen, N.U.N. | | Deposit date: | 2024-01-27 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Identification of FDA-approved drugs that induce heart regeneration in mammals.
Nat Cardiovasc Res, 3, 2024
|
|
4U13
 
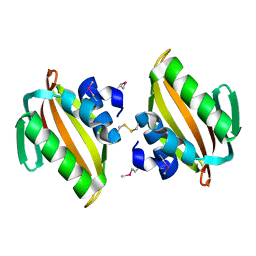 | | Crystal structure of putative polyketide cyclase (protein SMa1630) from Sinorhizobium meliloti at 2.3 A resolution | | Descriptor: | putative polyketide cyclase SMa1630 | | Authors: | Shabalin, I.G, Bacal, P, Osinski, T, Cooper, D.R, Szlachta, K, Stead, M, Grabowski, M, Hammonds, J, Ahmed, M, Hillerich, B.S, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-07-14 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative polyketide cyclase (protein SMa1630) from Sinorhizobium meliloti at 2.3 A resolution
to be published
|
|
4UC0
 
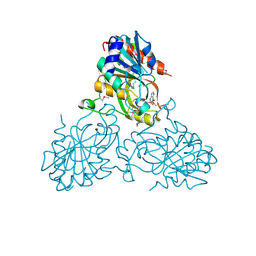 | | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis | | Descriptor: | HYPOXANTHINE, Purine nucleoside phosphorylase | | Authors: | Cameron, S.A, Sampathkumar, P, Ramagopal, U.A, Attonito, J, Ahmed, M, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Glenn, A.S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Wasserman, S.R, Schramm, V.L, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis
To be published
|
|
4REK
 
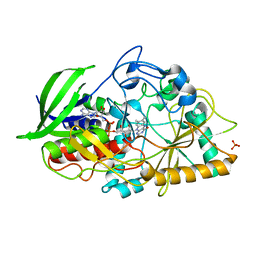 | | Crystal structure and charge density studies of cholesterol oxidase from Brevibacterium sterolicum at 0.74 ultra-high resolution | | Descriptor: | Cholesterol oxidase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Zarychta, B, Lyubimov, A, Ahmed, M, Munshi, P, Guillot, B, Vrielink, A. | | Deposit date: | 2014-09-23 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.74 Å) | | Cite: | Cholesterol oxidase: ultrahigh-resolution crystal structure and multipolar atom model-based analysis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7ATS
 
 | | The LIMK1 Kinase Domain Bound To LIJTF500127 | | Descriptor: | LIM domain kinase 1, N-[3-[5-(4-Chlorophenyl)-1H-pyrrolo[2,3-b]pyridine-3-carbonyl]-2,4-difluorophenyl]benzenesulfonamide | | Authors: | Mathea, S, Chatterjee, D, Preuss, F, Yamamoto, S, Tawada, M, Nomura, I, Takagi, T, Ahmed, M, Little, W, Mueller-Knapp, S, Knapp, S. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The LIMK1 Kinase Domain Bound To LIJTF500127
To Be Published
|
|
7ATU
 
 | | The LIMK1 Kinase Domain Bound To LIJTF500025 | | Descriptor: | (S)-2-benzyl-6-(8-chloro-5-methyl-4-oxo-2,3,4,5-tetrahydrobenzo[b][1,4]oxazepin-3-yl)-7-oxo-4,5,6,7-tetrahydro-2H-pyrazolo[3,4-c]pyridine-3-carboxamide, LIM domain kinase 1 | | Authors: | Mathea, S, Chatterjee, D, Preuss, F, Yamamoto, S, Tawada, M, Nomura, I, Takagi, T, Ahmed, M, Little, W, Mueller-Knapp, S, Knapp, S. | | Deposit date: | 2020-10-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The LIMK1 Kinase Domain Bound To LIJTF500025
To Be Published
|
|
4WED
 
 | | Crystal structure of ABC transporter substrate-binding protein from Sinorhizobium meliloti | | Descriptor: | ABC transporter, periplasmic solute-binding protein, FORMIC ACID, ... | | Authors: | Shabalin, I.G, Otwinowski, Z, Bacal, P, Cymborowski, M.T, Handing, K.B, Stead, M, Hammonds, J, Ahmed, M, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of ABC transporter substrate-binding protein from Sinorhizobium meliloti
to be published
|
|
4WBT
 
 | | Crystal structure of histidinol-phosphate aminotransferase from Sinorhizobium meliloti in complex with pyridoxal-5'-phosphate | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Shabalin, I.G, Bacal, P, Kowalska, A.K, Cooper, D.R, Stead, M, Hammonds, J, Ahmed, M, Hillerich, B.S, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-03 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of histidinol-phosphate aminotransferase from Sinorhizobium meliloti in complex with pyridoxal-5'-phosphate
to be published
|
|
4WJI
 
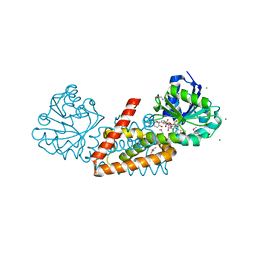 | | Crystal structure of cyclohexadienyl dehydrogenase from Sinorhizobium meliloti in complex with NADP and tyrosine | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Shabalin, I.G, Cooper, D.R, Hou, J, Zimmerman, M.D, Stead, M, Hillerich, B.S, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-30 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of cyclohexadienyl dehydrogenase from Sinorhizobium meliloti in complex with NADP
to be published
|
|
2BBR
 
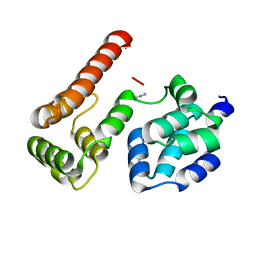 | | Crystal Structure of MC159 Reveals Molecular Mechanism of DISC Assembly and vFLIP Inhibition | | Descriptor: | AZIDE ION, Viral CASP8 and FADD-like apoptosis regulator | | Authors: | Yang, J.K, Wang, L, Zheng, L, Wan, F, Ahmed, M, Lenardo, M.J, Wu, H. | | Deposit date: | 2005-10-17 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition.
Mol.Cell, 20, 2005
|
|
2BBZ
 
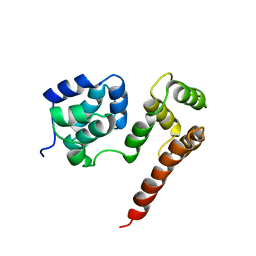 | | Crystal Structure of MC159 Reveals Molecular Mechanism of DISC Assembly and vFLIP Inhibition | | Descriptor: | Viral CASP8 and FADD-like apoptosis regulator | | Authors: | Yang, J.K, Wang, L, Zheng, L, Wan, F, Ahmed, M, Lenardo, M.J, Wu, H. | | Deposit date: | 2005-10-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition.
Mol.Cell, 20, 2005
|
|
5ZUR
 
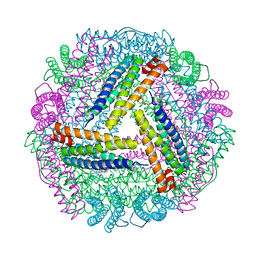 | | Achromobacter Dh1f Bacterioferritin | | Descriptor: | BARIUM ION, Bacterioferritin, CHLORIDE ION, ... | | Authors: | Dwivedy, A, Jha, B, Singh, K.H, Ahmed, M, Ashraf, A, Kumar, D, Biswal, B.K. | | Deposit date: | 2018-05-08 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Serendipitous crystallization and structure determination of bacterioferritin from Achromobacter.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5BXY
 
 | | Crystal structure of RNA methyltransferase from Salinibacter ruber in complex with S-Adenosyl-L-homocysteine | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, RNA methyltransferase, ... | | Authors: | Handing, K.B, LaRowe, C, Shabalin, I.G, Stead, M, Hillerich, B.S, Ahmed, M, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-09 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of RNA methylase family protein from Salinibacterruber in complex with S-Adenosyl-L-homocysteine.
to be published
|
|
5C9G
 
 | | Crystal Structure of a Putative enoyl-CoA hydratase/isomerase family protein from Hyphomonas neptunium | | Descriptor: | D-MALATE, Enoyl-CoA hydratase/isomerase family protein, TETRAETHYLENE GLYCOL | | Authors: | Szlachta, K, Cooper, D.R, Chapman, H.C, Cymbrowski, M.T, Stead, M, Hillerich, B.S, Ahmed, M, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, Hammonds, J, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a Putative enoyl-CoA hydratase/isomerase family protein from Hyphomonas neptunium
to be published
|
|
2M4M
 
 | | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata | | Descriptor: | hypothetical protein | | Authors: | Harris, R, Hillerich, B, Ahmed, M, Bonanno, J.B, Chamala, S, Evans, B, Lafleur, J, Hammonds, J, Washington, E, Stead, M, Love, J, Attonito, J, Patel, H, Seidel, R.D, Chook, Y.M, Rout, M.P, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2013-02-07 | | Release date: | 2013-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata
To be Published
|
|
2M4N
 
 | | Solution structure of the putative Ras interaction domain of AFD-1, isoform a from Caenorhabditis elegans | | Descriptor: | Protein AFD-1, isoform a | | Authors: | Harris, R, Hillerich, B, Ahmed, M, Bonanno, J.B, Chamala, S, Evans, B, Lafleur, J, Hammonds, J, Washington, E, Stead, M, Love, J, Attonito, J, Seidel, R.D, Liddington, R.C, Weis, W.I, Nelson, W.J, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Assembly, Dynamics and Evolution of Cell-Cell and Cell-Matrix Adhesions (CELLMAT) | | Deposit date: | 2013-02-07 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the putative Ras interaction domain of AFD-1, isoform a from Caenorhabditis elegans
To be Published
|
|
2M72
 
 | | Solution structure of uncharacterized thioredoxin-like protein PG_2175 from Porphyromonas gingivalis | | Descriptor: | Uncharacterized thioredoxin-like protein | | Authors: | Harris, R, Ahmed, M, Attonito, J, Bonanno, J.B, Chamala, S, Chowdhury, S, Evans, B, Fiser, A, Glenn, A.S, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Love, J.D, Seidel, R.D, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-16 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of uncharacterized thioredoxin-like protein PG_2175 from Porphyromonas gingivalis
To be Published
|
|
