1QUV
 
 | |
1APA
 
 | | X-RAY STRUCTURE OF A POKEWEED ANTIVIRAL PROTEIN, CODED BY A NEW GENOMIC CLONE, AT 0.23 NM RESOLUTION. A MODEL STRUCTURE PROVIDES A SUITABLE ELECTROSTATIC FIELD FOR SUBSTRATE BINDING. | | Descriptor: | POKEWEED ANTIVIRAL PROTEIN | | Authors: | Ago, H, Kataoka, J, Tsuge, H, Habuka, N, Inagaki, E, Noma, M, Miyano, M. | | Deposit date: | 1993-09-21 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of a pokeweed antiviral protein, coded by a new genomic clone, at 0.23 nm resolution. A model structure provides a suitable electrostatic field for substrate binding.
Eur.J.Biochem., 225, 1994
|
|
4YUU
 
 | | Crystal structure of oxygen-evolving photosystem II from a red alga | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ago, H, Shen, J.-R. | | Deposit date: | 2015-03-19 | | Release date: | 2016-01-20 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (2.7700038 Å) | | Cite: | Novel Features of Eukaryotic Photosystem II Revealed by Its Crystal Structure Analysis from a Red Alga
J.Biol.Chem., 291, 2016
|
|
1HAK
 
 | | CRYSTAL STRUCTURE OF RECOMBINANT HUMAN PLACENTAL ANNEXIN V COMPLEXED WITH K-201 AS A CALCIUM CHANNEL ACTIVITY INHIBITOR | | Descriptor: | 4-[3-{1-(4-BENZYL)PIPERODINYL}PROPIONYL]-7-METHOXY-2,3,4,5-TERTRAHYDRO-1,4-BENZOTHIAZEPINE, ANNEXIN V | | Authors: | Ago, H, Inagaki, E, Miyano, M. | | Deposit date: | 1997-12-10 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of annexin V with its ligand K-201 as a calcium channel activity inhibitor.
J.Mol.Biol., 274, 1997
|
|
2DDT
 
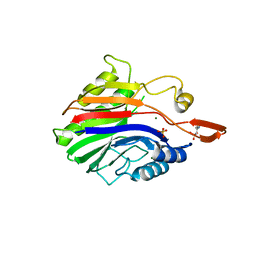 | | Crystal structure of sphingomyelinase from Bacillus cereus with magnesium ion | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Ago, H, Oda, M, Tsuge, H, Katunuma, N, Miyano, M, Sakurai, J, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-02 | | Release date: | 2006-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the sphingomyelin phosphodiesterase activity in neutral sphingomyelinase from Bacillus cereus.
J.Biol.Chem., 281, 2006
|
|
2DDS
 
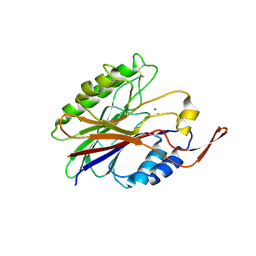 | | Crystal structure of sphingomyelinase from Bacillus cereus with cobalt ion | | Descriptor: | COBALT (II) ION, Sphingomyelin phosphodiesterase | | Authors: | Ago, H, Oda, M, Takahashi, M, Tsuge, H, Ochi, S, Katunuma, N, Miyano, M, Sakurai, J, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-02 | | Release date: | 2006-05-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Sphingomyelin Phosphodiesterase Activity in Neutral Sphingomyelinase from Bacillus cereus.
J.Biol.Chem., 281, 2006
|
|
2DDR
 
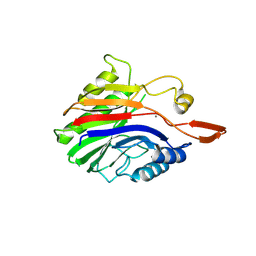 | | Crystal structure of sphingomyelinase from Bacillus cereus with calcium ion | | Descriptor: | CALCIUM ION, Sphingomyelin phosphodiesterase | | Authors: | Ago, H, Oda, M, Takahashi, M, Tsuge, H, Ochi, S, Katunuma, N, Miyano, M, Sakurai, J, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-02 | | Release date: | 2006-05-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of the Sphingomyelin Phosphodiesterase Activity in Neutral Sphingomyelinase from Bacillus cereus.
J.Biol.Chem., 281, 2006
|
|
1ULU
 
 | | Crystal structure of tt0143 from Thermus thermophilus HB8 | | Descriptor: | enoyl-acyl carrier protein reductase | | Authors: | Ago, H, Hamada, K, Ida, K, Kanda, H, Sugahara, M, Yamamoto, M, Nodake, Y, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of tt0143 from Thermus thermophilus HB8
To be Published
|
|
1ULQ
 
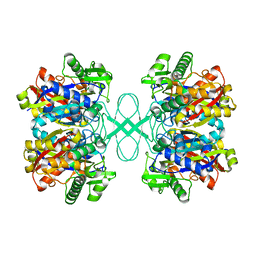 | | Crystal structure of tt0182 from Thermus thermophilus HB8 | | Descriptor: | putative acetyl-CoA acetyltransferase | | Authors: | Ago, H, Hamada, K, Ida, K, Kanda, H, Sugahara, M, Yamamoto, M, Kuroishi, C, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of tt0182 from Thermus thermophilus HB8
To be Published
|
|
2PNO
 
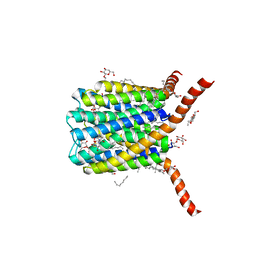 | | Crystal structure of human leukotriene C4 synthase | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, GLUTATHIONE, Leukotriene C4 synthase | | Authors: | Ago, H, Kanaoka, Y, Irikura, D, Lam, B.K, Shimamura, T, Austen, K.F, Miyano, M. | | Deposit date: | 2007-04-24 | | Release date: | 2007-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of a human membrane protein involved in cysteinyl leukotriene biosynthesis
Nature, 448, 2007
|
|
1ULR
 
 | | Crystal structure of tt0497 from Thermus thermophilus HB8 | | Descriptor: | putative acylphosphatase | | Authors: | Ago, H, Hamada, K, Sugahara, M, Kuroishi, C, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of tt0497 from Thermus thermophilus HB8
To be Published
|
|
8ILL
 
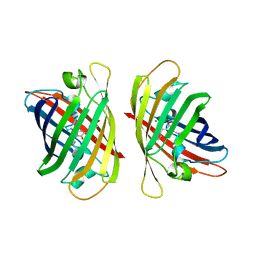 | | Crystal structure of a highly photostable and bright green fluorescent protein at pH5.6 | | Descriptor: | CHLORIDE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, green fluorescent protein | | Authors: | Ago, H, Ando, R, Hirano, M, Shimozono, S, Miyawaki, A, Yamamoto, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | StayGold variants for molecular fusion and membrane-targeting applications.
Nat.Methods, 21, 2024
|
|
8ILK
 
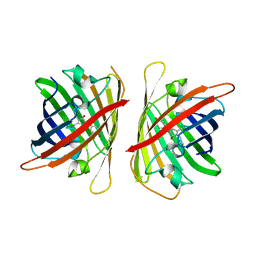 | | Crystal structure of a highly photostable and bright green fluorescent protein at pH8.5 | | Descriptor: | CHLORIDE ION, Green FLUORESCENT PROTEIN | | Authors: | Ago, H, Ando, R, Hirano, M, Shimozono, S, Miyawaki, A, Yamamoto, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | StayGold variants for molecular fusion and membrane-targeting applications.
Nat.Methods, 21, 2024
|
|
3WDR
 
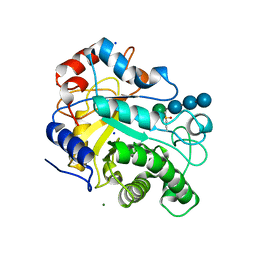 | | Crystal structure of beta-mannanase from a symbiotic protist of the termite Reticulitermes speratus complexed with gluco-manno-oligosaccharide | | Descriptor: | BICARBONATE ION, Beta-mannanase, MAGNESIUM ION, ... | | Authors: | Tsukagoshi, H, Ishida, T, Touhara, K.K, Igarashi, K, Samejima, M, Fushinobu, S, Kitamoto, K, Arioka, M. | | Deposit date: | 2013-06-20 | | Release date: | 2014-03-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biochemical Analyses of Glycoside Hydrolase Family 26 beta-Mannanase from a Symbiotic Protist of the Termite Reticulitermes speratus
J.Biol.Chem., 289, 2014
|
|
3WDQ
 
 | | Crystal structure of beta-mannanase from a symbiotic protist of the termite Reticulitermes speratus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-mannanase, MAGNESIUM ION, ... | | Authors: | Tsukagoshi, H, Ishida, T, Touhara, K.K, Igarashi, K, Samejima, M, Fushinobu, S, Kitamoto, K, Arioka, M. | | Deposit date: | 2013-06-20 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural and Biochemical Analyses of Glycoside Hydrolase Family 26 beta-Mannanase from a Symbiotic Protist of the Termite Reticulitermes speratus
J.Biol.Chem., 289, 2014
|
|
7Y44
 
 | | Re-refinement of damage free X-ray structure of bovine cytochrome c oxidase at 1.9 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Hirata, K, Ago, H. | | Deposit date: | 2022-06-14 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of damage-free crystal structure of an X-ray-sensitive protein using an XFEL.
Nat.Methods, 11, 2014
|
|
3B29
 
 | | Human leukotriene C4 synthase in complex with dodecyl-beta-D-selenomaltoside | | Descriptor: | GLUTATHIONE, Leukotriene C4 synthase, SULFATE ION, ... | | Authors: | Saino, H, Ago, H, Miyano, M. | | Deposit date: | 2011-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Seleno-detergent MAD phasing of leukotriene C4 synthase in complex with dodecyl-beta-D-selenomaltoside
Acta Crystallogr.,Sect.F, 67, 2011
|
|
5ONY
 
 | | As-isolated resting state copper nitrite reductase from Achromobacter xylosoxidans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Halsted, T.P, Yamashita, K, Hirata, K, Ago, H, Ueno, G, Tosha, T, Eady, R.R, Antonyuk, S.V, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An unprecedented dioxygen species revealed by serial femtosecond rotation crystallography in copper nitrite reductase.
IUCrJ, 5, 2018
|
|
5ONX
 
 | | Resting state copper nitrite reductase determined by serial femtosecond rotation crystallography | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Halsted, T.P, Yamashita, K, Hirata, K, Ago, H, Ueno, G, Tosha, T, Eady, R.R, Antonyuk, S.V, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An unprecedented dioxygen species revealed by serial femtosecond rotation crystallography in copper nitrite reductase.
IUCrJ, 5, 2018
|
|
5X1B
 
 | | CO bound cytochrome c oxidase at 20 nsec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
5X19
 
 | | CO bound cytochrome c oxidase at 100 micro sec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
8J81
 
 | | MDM2 bound with a peptoid | | Descriptor: | (2S)-2-[[(2S)-2-[(6-chloranyl-1H-indol-3-yl)methyl-[(2S)-2-[[(2S)-2-[ethanoyl-(phenylmethyl)amino]propanoyl]-methyl-amino]propanoyl]amino]propanoyl]-methyl-amino]-N-(3,3-dimethylbutyl)-N-[(2S)-1-oxidanylidene-1-piperazin-1-yl-propan-2-yl]propanamide, CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Yokomine, M, Fukuda, Y, Ago, H, Matsuura, H, Ueno, G, Nagatoishi, S, Yamamoto, M, Tsumoto, K, Jumpei, M, Sando, S. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A structural and physicochemical study of how a peptoid binds to a protein
To Be Published
|
|
4UB6
 
 | | Native structure of photosystem II (dataset-1) by a femtosecond X-ray laser | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Akita, F, Hirata, K, Ueno, G, Murakami, H, Nakajima, Y, Shimizu, T, Yamashita, K, Yamamoto, M, Ago, H, Shen, J.R. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Native structure of photosystem II at 1.95 angstrom resolution viewed by femtosecond X-ray pulses.
Nature, 517, 2015
|
|
4UB8
 
 | | Native structure of photosystem II (dataset-2) by a femtosecond X-ray laser | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Akita, F, Hirata, K, Ueno, G, Murakami, H, Nakajima, Y, Shimizu, T, Yamashita, K, Yamamoto, M, Ago, H, Shen, J.R. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Native structure of photosystem II at 1.95 angstrom resolution viewed by femtosecond X-ray pulses.
Nature, 517, 2015
|
|
3PCV
 
 | | Crystal structure analysis of human leukotriene C4 synthase at 1.9 angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, DODECYL-BETA-D-MALTOSIDE, GLUTATHIONE, ... | | Authors: | Saino, H, Ago, H, Miyano, M. | | Deposit date: | 2010-10-22 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The catalytic architecture of leukotriene C4 synthase with two arginine residues
J.Biol.Chem., 286, 2011
|
|
