2OUA
 
 | | Crystal Structure of Nocardiopsis Protease (NAPase) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, GLYCEROL, ... | | Authors: | Kelch, B.A, Agard, D.A. | | Deposit date: | 2007-02-09 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and mechanistic exploration of Acid resistance: kinetic stability facilitates evolution of extremophilic behavior
J.Mol.Biol., 368, 2007
|
|
2PFE
 
 | | Crystal Structure of Thermobifida fusca Protease A (TFPA) | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, Alkaline serine protease, GLYCEROL, ... | | Authors: | Kelch, B.A, Agard, D.A. | | Deposit date: | 2007-04-04 | | Release date: | 2007-07-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.436 Å) | | Cite: | Mesophile versus Thermophile: Insights Into the Structural Mechanisms of Kinetic Stability
J.Mol.Biol., 370, 2007
|
|
2PRO
 
 | |
1K7B
 
 | | NMR Solution Structure of sTva47, the Viral-Binding Domain of Tva | | Descriptor: | SUBGROUP A ROUS SARCOMA VIRUS RECEPTOR PG800 AND PG950 | | Authors: | Tonelli, M, Peters, R.J, James, T.L, Agard, D.A. | | Deposit date: | 2001-10-18 | | Release date: | 2001-12-19 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the viral binding domain of Tva, the cellular receptor for subgroup A avian leukosis and sarcoma virus.
FEBS Lett., 509, 2001
|
|
1L2J
 
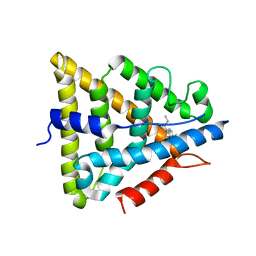 | | Human Estrogen Receptor beta Ligand-binding Domain in Complex with (R,R)-5,11-cis-diethyl-5,6,11,12-tetrahydrochrysene-2,8-diol | | Descriptor: | (R,R)-5,11-CIS-DIETHYL-5,6,11,12-TETRAHYDROCHRYSENE-2,8-DIOL, ESTROGEN RECEPTOR BETA | | Authors: | Shiau, A.K, Barstad, D, Radek, J.T, Meyers, M.J, Nettles, K.W, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Agard, D.A, Greene, G.L. | | Deposit date: | 2002-02-21 | | Release date: | 2002-05-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural characterization of a subtype-selective ligand reveals a novel mode of estrogen receptor antagonism.
Nat.Struct.Biol., 9, 2002
|
|
1P11
 
 | |
1P12
 
 | |
1QRX
 
 | |
1QRW
 
 | | CRYSTAL STRUCTURE OF AN ALPHA-LYTIC PROTEASE MUTANT WITH ACCELERATED FOLDING KINETICS, R102H/G134S, PH 8 | | Descriptor: | ALHPA-LYTIC PROTEASE, GLYCEROL, SULFATE ION | | Authors: | Derman, A.I, Mau, T, Agard, D.A. | | Deposit date: | 1999-06-16 | | Release date: | 1999-06-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Genetic Screen That Targets Specifically the Folding Transition State of Alpha-Lytic Protease
To be Published, 1999
|
|
7KCK
 
 | |
7KCL
 
 | |
7KCM
 
 | |
7KLU
 
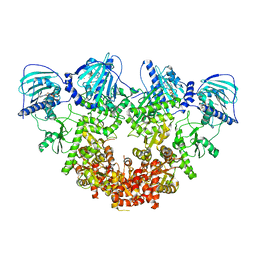 | |
7KLV
 
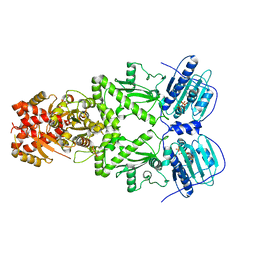 | |
7KW7
 
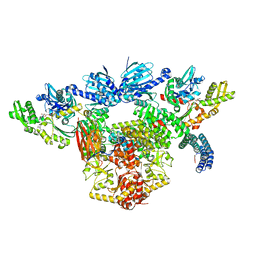 | | Atomic cryoEM structure of Hsp90-Hsp70-Hop-GR | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glucocorticoid receptor, Heat shock 70 kDa protein 1A, ... | | Authors: | Wang, R.Y, Noddings, C.M, Kirschke, E, Myasnikov, A, Johnson, J.L, Agard, D.A. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure of Hsp90-Hsp70-Hop-GR reveals the Hsp90 client-loading mechanism.
Nature, 601, 2022
|
|
7KRJ
 
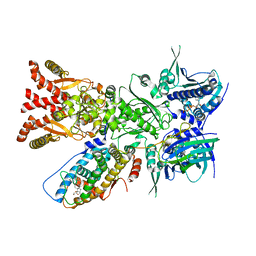 | | The GR-Maturation Complex: Glucocorticoid Receptor in complex with Hsp90 and co-chaperone p23 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DEXAMETHASONE, Glucocorticoid receptor, ... | | Authors: | Noddings, C.M, Wang, Y.-R, Agard, D.A. | | Deposit date: | 2020-11-20 | | Release date: | 2022-01-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structure of Hsp90-p23-GR reveals the Hsp90 client-remodelling mechanism.
Nature, 601, 2021
|
|
7KOD
 
 | | Cryo-EM structure of heavy chain mouse apoferritin | | Descriptor: | Ferritin heavy chain | | Authors: | Sun, M, Azumaya, C, Tse, E, Frost, A, Southworth, D, Verba, K.A, Cheng, Y, Agard, D.A. | | Deposit date: | 2020-11-08 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.655 Å) | | Cite: | Practical considerations for using K3 cameras in CDS mode for high-resolution and high-throughput single particle cryo-EM.
J.Struct.Biol., 213, 2021
|
|
