3J40
 
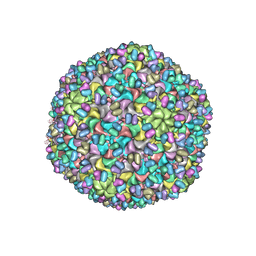 | | Validated Near-Atomic Resolution Structure of Bacteriophage Epsilon15 Derived from Cryo-EM and Modeling | | Descriptor: | gp10, gp7 | | Authors: | Baker, M.L, Hryc, C.F, Zhang, Q, Wu, W, Jakana, J, Haase-Pettingell, C, Afonine, P.V, Adams, P.D, King, J.A, Jiang, W, Chiu, W. | | Deposit date: | 2013-05-30 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Validated near-atomic resolution structure of bacteriophage epsilon15 derived from cryo-EM and modeling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3J7L
 
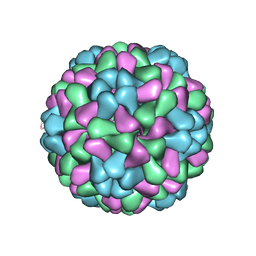 | | Full virus map of brome mosaic virus | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3J7N
 
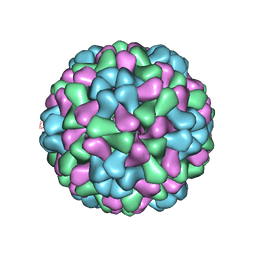 | | Virus model of brome mosaic virus (second half data set) | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3J7M
 
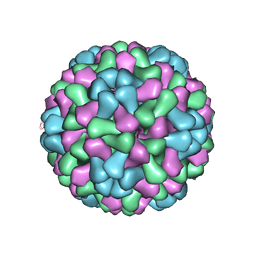 | | Virus model of brome mosaic virus (first half data set) | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3OS9
 
 | | Estrogen Receptor | | Descriptor: | 4-[1-allyl-7-(trifluoromethyl)-1H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor | | Authors: | Bruning, J, Parent, A.A, Gil, G, Zhao, M, Nowak, J, Pace, M.C, Smith, C.L, Afonine, P.V, Adams, P.D, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2010-09-08 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Coupling of receptor conformation and ligand orientation determine graded activity.
Nat.Chem.Biol., 6, 2010
|
|
4OW3
 
 | | Thermolysin structure determined by free-electron laser | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Hattne, J, Echols, N, Tran, R, Kern, J, Gildea, R.J, Brewster, A.S, Alonso-Mori, R, Glockner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, White, W.E, Schafer, D.W, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Glatzel, P, Zwart, P.H, Grosse-Kunstleve, R.W, Bogan, M.J, Messerschmidt, M, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Yano, J, Bergmann, U, Yachandra, V.K, Adams, P.D, Sauter, N.K. | | Deposit date: | 2014-01-30 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Accurate macromolecular structures using minimal measurements from X-ray free-electron lasers.
Nat.Methods, 11, 2014
|
|
7T1J
 
 | | Crystal structure of RUBISCO from Rhodospirillaceae bacterium BRH_c57 | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Pereira, J.H, Liu, A.K, Shih, P.M, Adams, P.D. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural plasticity enables evolution and innovation of RuBisCO assemblies.
Sci Adv, 8, 2022
|
|
5J54
 
 | | The Structure and Mechanism of NOV1, a Resveratrol-Cleaving Dioxygenase | | Descriptor: | Carotenoid oxygenase, FE (III) ION, OXYGEN MOLECULE, ... | | Authors: | McAndrew, R.P, Pereira, J.H, Sathitsuksanoh, N, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2016-04-01 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
8D3Z
 
 | | Crystal structure of GalS1 in complex with Manganese from Populus trichocarpas | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Galactan synthase, ... | | Authors: | Pereira, J.H, Prabhakar, P.K, Urbanowicz, B.R, Adams, P.D. | | Deposit date: | 2022-06-01 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural and biochemical insight into a modular beta-1,4-galactan synthase in plants.
Nat.Plants, 9, 2023
|
|
8D3T
 
 | | Crystal structure of GalS1 from Populus trichocarpas | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pereira, J.H, Prabhakar, P.K, Urbanowicz, B.R, Adams, P.D. | | Deposit date: | 2022-06-01 | | Release date: | 2023-03-15 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural and biochemical insight into a modular beta-1,4-galactan synthase in plants.
Nat.Plants, 9, 2023
|
|
5J53
 
 | | The Structure and Mechanism of NOV1, a Resveratrol-Cleaving Dioxygenase | | Descriptor: | Carotenoid oxygenase, FE (III) ION, OXYGEN MOLECULE, ... | | Authors: | McAndrew, R.P, Pereira, J.H, Sathitsuksanoh, N, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2016-04-01 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
7RF6
 
 | | RT XFEL structure of Photosystem II 250 microseconds after the second illumination at 2.01 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Hussein, R, Ibrahim, M, Bhowmick, A, Simon, P.S, Chatterjee, R, Lassalle, L, Doyle, M.D, Bogacz, I, Kim, I.-S, Cheah, M.H, Gul, S, de Lichtenberg, C, Chernev, P, Pham, C.C, Young, I.D, Carbajo, S, Fuller, F.D, Alonso-Mori, R, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bolotovski, R, Mendez, D, Holton, J.M, Moriarty, N.W, Adams, P.D, Bergmann, U, Sauter, N.K, Dobbek, H, Messinger, J, Zouni, A, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2021-07-13 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural dynamics in the water and proton channels of photosystem II during the S 2 to S 3 transition.
Nat Commun, 12, 2021
|
|
7RF4
 
 | | RT XFEL structure of Photosystem II 50 microseconds after the second illumination at 2.27 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Hussein, R, Ibrahim, M, Bhowmick, A, Simon, P.S, Chatterjee, R, Lassalle, L, Doyle, M.D, Bogacz, I, Kim, I.-S, Cheah, M.H, Gul, S, de Lichtenberg, C, Chernev, P, Pham, C.C, Young, I.D, Carbajo, S, Fuller, F.D, Alonso-Mori, R, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bolotovski, R, Mendez, D, Holton, J.M, Moriarty, N.W, Adams, P.D, Bergmann, U, Sauter, N.K, Dobbek, H, Messinger, J, Zouni, A, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2021-07-13 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural dynamics in the water and proton channels of photosystem II during the S 2 to S 3 transition.
Nat Commun, 12, 2021
|
|
7RF2
 
 | | RT XFEL structure of dark-stable state of Photosystem II (0F, S1 rich) at 2.08 Angstrom | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Hussein, R, Ibrahim, M, Bhowmick, A, Simon, P.S, Chatterjee, R, Lassalle, L, Doyle, M.D, Bogacz, I, Kim, I.-S, Cheah, M.H, Gul, S, de Lichtenberg, C, Chernev, P, Pham, C.C, Young, I.D, Carbajo, S, Fuller, F.D, Alonso-Mori, R, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bolotovski, R, Mendez, D, Holton, J.M, Moriarty, N.W, Adams, P.D, Bergmann, U, Sauter, N.K, Dobbek, H, Messinger, J, Zouni, A, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2021-07-13 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural dynamics in the water and proton channels of photosystem II during the S 2 to S 3 transition.
Nat Commun, 12, 2021
|
|
7RF8
 
 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Hussein, R, Ibrahim, M, Bhowmick, A, Simon, P.S, Chatterjee, R, Lassalle, L, Doyle, M.D, Bogacz, I, Kim, I.-S, Cheah, M.H, Gul, S, de Lichtenberg, C, Chernev, P, Pham, C.C, Young, I.D, Carbajo, S, Fuller, F.D, Alonso-Mori, R, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bolotovski, R, Mendez, D, Holton, J.M, Moriarty, N.W, Adams, P.D, Bergmann, U, Sauter, N.K, Dobbek, H, Messinger, J, Zouni, A, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2021-07-13 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural dynamics in the water and proton channels of photosystem II during the S 2 to S 3 transition.
Nat Commun, 12, 2021
|
|
7RF3
 
 | | RT XFEL structure of the one-flash state of Photosystem II (1F, S2-rich) at 2.26 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Hussein, R, Ibrahim, M, Bhowmick, A, Simon, P.S, Chatterjee, R, Lassalle, L, Doyle, M.D, Bogacz, I, Kim, I.-S, Cheah, M.H, Gul, S, de Lichtenberg, C, Chernev, P, Pham, C.C, Young, I.D, Carbajo, S, Fuller, F.D, Alonso-Mori, R, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bolotovski, R, Mendez, D, Holton, J.M, Moriarty, N.W, Adams, P.D, Bergmann, U, Sauter, N.K, Dobbek, H, Messinger, J, Zouni, A, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2021-07-13 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural dynamics in the water and proton channels of photosystem II during the S 2 to S 3 transition.
Nat Commun, 12, 2021
|
|
7RF7
 
 | | RT XFEL structure of Photosystem II 400 microseconds after the second illumination at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Hussein, R, Ibrahim, M, Bhowmick, A, Simon, P.S, Chatterjee, R, Lassalle, L, Doyle, M.D, Bogacz, I, Kim, I.-S, Cheah, M.H, Gul, S, de Lichtenberg, C, Chernev, P, Pham, C.C, Young, I.D, Carbajo, S, Fuller, F.D, Alonso-Mori, R, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bolotovski, R, Mendez, D, Holton, J.M, Moriarty, N.W, Adams, P.D, Bergmann, U, Sauter, N.K, Dobbek, H, Messinger, J, Zouni, A, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2021-07-13 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural dynamics in the water and proton channels of photosystem II during the S 2 to S 3 transition.
Nat Commun, 12, 2021
|
|
7RF5
 
 | | RT XFEL structure of Photosystem II 150 microseconds after the second illumination at 2.23 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Hussein, R, Ibrahim, M, Bhowmick, A, Simon, P.S, Chatterjee, R, Lassalle, L, Doyle, M.D, Bogacz, I, Kim, I.-S, Cheah, M.H, Gul, S, de Lichtenberg, C, Chernev, P, Pham, C.C, Young, I.D, Carbajo, S, Fuller, F.D, Alonso-Mori, R, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bolotovski, R, Mendez, D, Holton, J.M, Moriarty, N.W, Adams, P.D, Bergmann, U, Sauter, N.K, Dobbek, H, Messinger, J, Zouni, A, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2021-07-13 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural dynamics in the water and proton channels of photosystem II during the S 2 to S 3 transition.
Nat Commun, 12, 2021
|
|
7RF1
 
 | | RT XFEL structure of Photosystem II averaged across all S-states at 1.89 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Hussein, R, Ibrahim, M, Bhowmick, A, Simon, P.S, Chatterjee, R, Lassalle, L, Doyle, M.D, Bogacz, I, Kim, I.-S, Cheah, M.H, Gul, S, de Lichtenberg, C, Chernev, P, Pham, C.C, Young, I.D, Carbajo, S, Fuller, F.D, Alonso-Mori, R, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bolotovski, R, Mendez, D, Holton, J.M, Moriarty, N.W, Adams, P.D, Bergmann, U, Sauter, N.K, Dobbek, H, Messinger, J, Zouni, A, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2021-07-13 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural dynamics in the water and proton channels of photosystem II during the S 2 to S 3 transition.
Nat Commun, 12, 2021
|
|
4OPI
 
 | | Constructing tailored isoprenoid products by structure-guided modification of geranylgeranyl reductase. | | Descriptor: | Conserved Archaeal protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GERANYLGERANYL DIPHOSPHATE | | Authors: | McAndrew, R.P, Kung, Y, Xie, X, Liu, C, Pereira, J.H, Keasling, J.D, Adams, P.D. | | Deposit date: | 2014-02-05 | | Release date: | 2014-07-09 | | Last modified: | 2014-07-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Constructing tailored isoprenoid products by structure-guided modification of geranylgeranyl reductase.
Structure, 22, 2014
|
|
4OPT
 
 | | Constructing tailored isoprenoid products by structure-guided modification of geranylgeranyl reductase | | Descriptor: | Conserved Archaeal protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GERANYLGERANYL DIPHOSPHATE, ... | | Authors: | McAndrew, R.P, Kung, Y, Xie, X, Liu, C, Pereira, J.H, Keasling, J.D, Adams, P.D. | | Deposit date: | 2014-02-06 | | Release date: | 2014-07-09 | | Last modified: | 2014-07-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Constructing tailored isoprenoid products by structure-guided modification of geranylgeranyl reductase.
Structure, 22, 2014
|
|
4FE1
 
 | | Improving the Accuracy of Macromolecular Structure Refinement at 7 A Resolution | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Fromme, R, Adams, P.D, Fromme, P, Levitt, M, Schroeder, G.F, Brunger, A.T. | | Deposit date: | 2012-05-29 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.9228 Å) | | Cite: | Improving the accuracy of macromolecular structure refinement at 7 A resolution.
Structure, 20, 2012
|
|
4OPC
 
 | | Constructing tailored isoprenoid products by structure-guided modification of geranylgeranyl reductase. | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Conserved Archaeal protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | Authors: | McAndrew, R.P, Kung, Y, Xie, X, Liu, C, Pereira, J.H, Keasling, J.D, Adams, P.D. | | Deposit date: | 2014-02-05 | | Release date: | 2014-07-09 | | Last modified: | 2014-07-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Constructing tailored isoprenoid products by structure-guided modification of geranylgeranyl reductase.
Structure, 22, 2014
|
|
4OPU
 
 | | Constructing tailored isoprenoid products by structure-guided modification of geranylgeranyl reductase | | Descriptor: | Conserved Archaeal protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GERANYLGERANYL DIPHOSPHATE | | Authors: | McAndrew, R.P, Kung, Y, Xie, X, Liu, C, Pereira, J.H, Keasling, J.D, Adams, P.D. | | Deposit date: | 2014-02-06 | | Release date: | 2014-07-09 | | Last modified: | 2014-07-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Constructing tailored isoprenoid products by structure-guided modification of geranylgeranyl reductase.
Structure, 22, 2014
|
|
5J55
 
 | | The Structure and Mechanism of NOV1, a Resveratrol-Cleaving Dioxygenase | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, Carotenoid oxygenase, FE (III) ION, ... | | Authors: | McAndrew, R.P, Pereira, J.H, Sathitsuksanoh, N, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2016-04-01 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
