4JPW
 
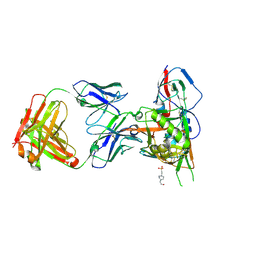 | | Crystal structure of broadly and potently neutralizing antibody 12a21 in complex with hiv-1 strain 93th057 gp120 mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEAVY CHAIN OF ANTIBODY 12A21, ... | | Authors: | Acharya, P, Luongo, T, Zhou, T, Kwong, P.D. | | Deposit date: | 2013-03-19 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization.
Cell(Cambridge,Mass.), 153, 2013
|
|
2FHJ
 
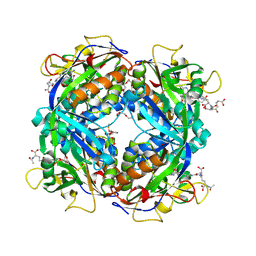 | | Crystal structure of formylmethanofuran: tetrahydromethanopterin formyltransferase in complex with its coenzymes | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, 5-(4-{[1-(2-AMINO-5-FORMYL-7-METHYL-4-OXO-3,4,5,6,7,8-HEXAHYDROPTERIDIN-6-YL)ETHYL]AMINO}PHENYL)-5-DEOXY-1-O-{5-O-[(1,3-DICARBOXYPROPOXY)(HYDROXY)PHOSPHORYL]PENTOFURANOSYL}PENTITOL, ... | | Authors: | Acharya, P, Warkentin, E, Thauer, R.K, Shima, S, Ermler, U. | | Deposit date: | 2005-12-25 | | Release date: | 2006-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of formylmethanofuran: tetrahydromethanopterin formyltransferase in complex with its coenzymes
J.Mol.Biol., 357, 2006
|
|
2FHK
 
 | | Crystal structure of formylmethanofuran: tetrahydromethanopterin formyltransferase in complex with its coenzymes | | Descriptor: | Formylmethanofuran--tetrahydromethanopterin formyltransferase, N-[4,5,7-TRICARBOXYHEPTANOYL]-L-GAMMA-GLUTAMYL-N-{2-[4-({5-[(FORMYLAMINO)METHYL]-3-FURYL}METHOXY)PHENYL]ETHYL}-D-GLUTAMINE, POTASSIUM ION | | Authors: | Acharya, P, Warkentin, E, Thauer, R.K, Shima, S, Ermler, U. | | Deposit date: | 2005-12-25 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of formylmethanofuran: tetrahydromethanopterin formyltransferase in complex with its coenzymes
J.Mol.Biol., 357, 2006
|
|
7SG4
 
 | |
8CSA
 
 | |
6XRJ
 
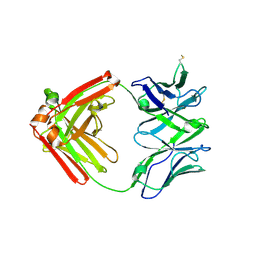 | |
8FEJ
 
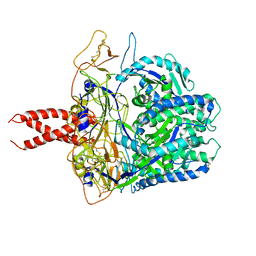 | |
8FEL
 
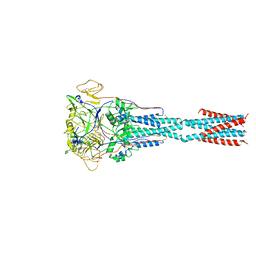 | |
8GDO
 
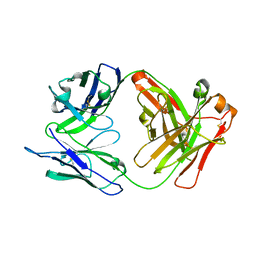 | |
8DTO
 
 | | Vaccine elicited Antibody MU89 bound to CH848.D949.10.17_N133D_N138T.DS.SOSIP.664 HIV-1 Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stalls, V, Acharya, P. | | Deposit date: | 2022-07-26 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Mutation-guided vaccine design: A process for developing boosting immunogens for HIV broadly neutralizing antibody induction.
Cell Host Microbe, 32, 2024
|
|
8DY6
 
 | | Vaccine elicited Antibody MU89+S27Y bound to CH848.D949.10.17_N133D_N138T.DS.SOSIP.664 HIV-1 Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stalls, V, Acharya, P. | | Deposit date: | 2022-08-03 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Mutation-guided vaccine design: A process for developing boosting immunogens for HIV broadly neutralizing antibody induction.
Cell Host Microbe, 32, 2024
|
|
8DKF
 
 | | Antibody DH1030.1 Fab fragment | | Descriptor: | DH1030.1 Heavy chain, DH1030.1 Light chain | | Authors: | Gobeil, S, Acharya, P. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | DH1030.1 glycan reactive Ab
To Be Published
|
|
8DP1
 
 | | Cryo-EM structure of HIV-1 Env(BG505.T332N SOSIP) in complex with DH1030.1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DH1030.1 Fab Heavy chain, ... | | Authors: | Gobeil, S, Acharya, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Shared recognition mechanism for HIV-1 envelope-reactive V3 glycan broadly neutralizing B cell lineage maturation in humans and macaques
To Be Published
|
|
8DOW
 
 | | Cryo-EM structure of HIV-1 Env(CH848 10.17 DS.SOSIP_DT) in complex with DH1030.1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DH1030.1 Fab Heavy chain, ... | | Authors: | Gobeil, S, Acharya, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Shared recognition mechanism for HIV-1 envelope-reactive V3 glycan broadly neutralizing B cell lineage maturation in humans and macaques
To Be Published
|
|
8DTK
 
 | |
6X2A
 
 | |
8DPZ
 
 | |
6X2B
 
 | |
6X29
 
 | |
6X2C
 
 | |
8EU8
 
 | | Cryo-EM structure of CH848 10.17DT DS-SOSIP-2P Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848 10.17DT SOSIP Envelope glycoprotein gp160 | | Authors: | Wrapp, D, Acharya, P, Haynes, B.F. | | Deposit date: | 2022-10-18 | | Release date: | 2023-01-04 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structure-Based Stabilization of SOSIP Env Enhances Recombinant Ectodomain Durability and Yield.
J.Virol., 97, 2023
|
|
6V8Z
 
 | |
6V8X
 
 | | VRC01 Bound BG505 F14 HIV-1 SOSIP Envelope Trimer Structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2019-12-12 | | Release date: | 2020-02-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Disruption of the HIV-1 Envelope allosteric network blocks CD4-induced rearrangements.
Nat Commun, 11, 2020
|
|
4H8W
 
 | | Crystal structure of non-neutralizing and ADCC-potent antibody N5-i5 in complex with HIV-1 clade A/E gp120 and sCD4. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB HEAVY CHAIN OF ADCC AND NON-NEUTRALIZING ANTI-HIV-1 ANTIBODY N5-I5, ... | | Authors: | Tolbert, W.D, Acharya, P, Kwong, P.D, Pazgier, M. | | Deposit date: | 2012-09-24 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Definition of an Antibody-Dependent Cellular Cytotoxicity Response Implicated in Reduced Risk for HIV-1 Infection.
J.Virol., 88, 2014
|
|
8UKD
 
 | |
