1GPQ
 
 | | Structure of ivy complexed with its target, HEWL | | Descriptor: | INHIBITOR OF VERTEBRATE LYSOZYME, LYSOZYME C | | Authors: | Abergel, C, Monchois, V, Claverie, J.-M. | | Deposit date: | 2001-11-08 | | Release date: | 2003-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Evolution of the Ivy Protein Family, Unexpected Lysozyme Inhibitors in Gram-Negative Bacteria.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1XS0
 
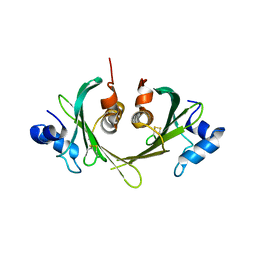 | | Structure of the E. coli Ivy protein | | Descriptor: | Inhibitor of vertebrate lysozyme | | Authors: | Abergel, C, Monchois, V, Byrn, D, Lazzaroni, J.C, Claverie, J.M. | | Deposit date: | 2004-10-18 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure and evolution of the Ivy protein family, unexpected lysozyme inhibitors in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1H1O
 
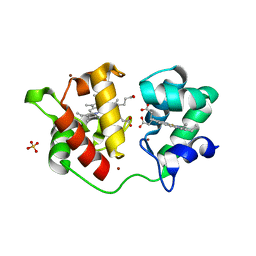 | | Acidithiobacillus ferrooxidans cytochrome c4 structure supports a complex-induced tuning of electron transfer | | Descriptor: | CYTOCHROME C-552, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Abergel, C, Nitschke, W, Malarte, G, Bruschi, M, Claverie, J.-M, Guidici-Orticoni, M.-T. | | Deposit date: | 2002-07-19 | | Release date: | 2003-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The Structure of Acidithiobacillus Ferrooxidans C(4)-Cytochrome. A Model for Complex-Induced Electron Transfer Tuning
Structure, 11, 2003
|
|
1CRZ
 
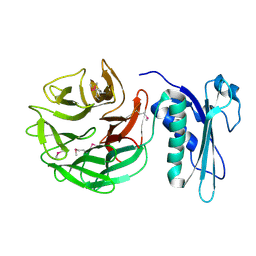 | | CRYSTAL STRUCTURE OF THE E. COLI TOLB PROTEIN | | Descriptor: | TOLB PROTEIN | | Authors: | Abergel, C, Bouveret, E, Claverie, J.-M, Brown, K, Rigal, A, Lazdunski, C, Benedetti, H. | | Deposit date: | 1999-08-16 | | Release date: | 2000-08-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Escherichia coli TolB protein determined by MAD methods at 1.95 A resolution.
Structure Fold.Des., 7, 1999
|
|
4BNQ
 
 | |
2J5B
 
 | | Structure of the Tyrosyl tRNA synthetase from Acanthamoeba polyphaga Mimivirus complexed with tyrosynol | | Descriptor: | 4-[(2S)-2-amino-3-hydroxypropyl]phenol, TYROSYL-TRNA SYNTHETASE | | Authors: | Abergel, C, Rudinger-thirion, J, Giege, R, Claverie, J.M. | | Deposit date: | 2006-09-13 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Virus-Encoded Aminoacyl-tRNA Synthetases: Structural and Functional Characterization of Mimivirus Tyrrs and Metrs.
J.Virol., 81, 2007
|
|
2G9Z
 
 | | Thiamin pyrophosphokinase from Candida albicans | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-5-(2-{[HYDROXY(PHOSPHONOAMINO)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-I UM, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Abergel, C, Santini, S, Monchois, V, Rousselle, T, Claverie, J.M, Bacterial targets at IGS-CNRS, France (BIGS) | | Deposit date: | 2006-03-07 | | Release date: | 2006-04-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural characterization of CA1462, the Candida albicans thiamine pyrophosphokinase.
Bmc Struct.Biol., 8, 2008
|
|
1OKJ
 
 | |
2HH9
 
 | | Thiamin pyrophosphokinase from Candida albicans | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, MAGNESIUM ION, Thiamin pyrophosphokinase | | Authors: | Abergel, C, Santini, S, Monchois, V, Rousselle, T, Claverie, J.M, Bacterial targets at IGS-CNRS, France (BIGS) | | Deposit date: | 2006-06-28 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of CA1462, the Candida albicans thiamine pyrophosphokinase.
Bmc Struct.Biol., 8, 2008
|
|
1MZR
 
 | | Structure of dkga from E.coli at 2.13 A resolution solved by molecular replacement | | Descriptor: | 2,5-diketo-D-gluconate reductase A, GLYCEROL, PHOSPHATE ION | | Authors: | Abergel, C, Jeudy, S, Monchois, V, Claverie, J.M, Bacterial targets at IGS-CNRS, France (BIGS) | | Deposit date: | 2002-10-09 | | Release date: | 2003-10-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of Escherichia coli DkgA, a broad-specificity aldo-keto reductase.
Proteins, 62, 2006
|
|
1UV0
 
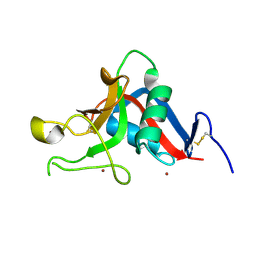 | | Pancreatitis-associated protein 1 from human | | Descriptor: | PANCREATITIS-ASSOCIATED PROTEIN 1, ZINC ION | | Authors: | Abergel, C, Shepard, W, Christal, L. | | Deposit date: | 2004-01-12 | | Release date: | 2004-01-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystallization and preliminary crystallographic study of HIP/PAP, a human C-lectin overexpressed in primary liver cancers.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1UUZ
 
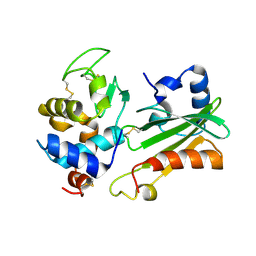 | | IVY:A NEW FAMILY OF PROTEIN | | Descriptor: | INHIBITOR OF VERTEBRATE LYSOZYME, LYSOZYME C | | Authors: | Abergel, C, Lembo, F, Byrne, D, Maza, C, Claverie, J.M. | | Deposit date: | 2004-01-12 | | Release date: | 2004-01-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Evolution of the Ivy Protein Family, Unexpected Lysozyme Inhibitors in Gram-Negative Bacteria.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1OAP
 
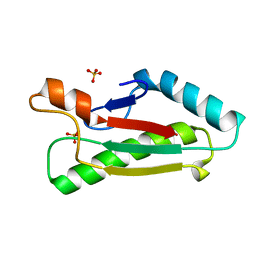 | | Mad structure of the periplasmique domain of the Escherichia coli PAL protein | | Descriptor: | PEPTIDOGLYCAN-ASSOCIATED LIPOPROTEIN, SULFATE ION | | Authors: | Abergel, C, Walburger, A, Bouveret, E, Claverie, J.M. | | Deposit date: | 2003-01-20 | | Release date: | 2004-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystallization and preliminary crystallographic study of the peptidoglycan-associated lipoprotein from Escherichia coli.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7PTV
 
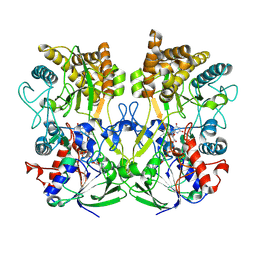 | | Structure of the Mimivirus genomic fibre asymmetric unit | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
7QRJ
 
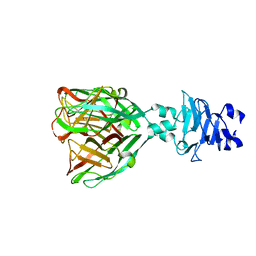 | | Crystal structure of Zamilon vitis protein Zav_19 | | Descriptor: | Zav_19 protein | | Authors: | Jeudy, S, Abergel, C. | | Deposit date: | 2022-01-11 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The fibre head structure used by unrelated families of viruses is unexpectedly a major component of the Marseilleviridae and Zamilon virophages capsids
To Be Published
|
|
7QRR
 
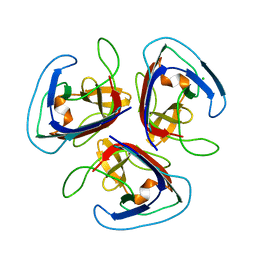 | | Crystal structure of Noumeavirus NMV_189 protein | | Descriptor: | CHLORIDE ION, NMV_189 protein, PHOSPHATE ION | | Authors: | Jeudy, S, Abergel, C. | | Deposit date: | 2022-01-11 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The fibre head structure used by unrelated families of viruses is unexpectedly a major component of the Marseilleviridae and Zamilon virophages capsids
To Be Published
|
|
7YX5
 
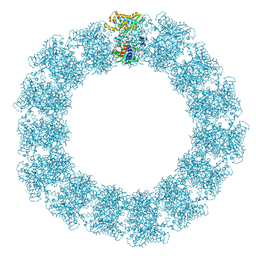 | | Structure of the Mimivirus genomic fibre in its relaxed 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
8ORH
 
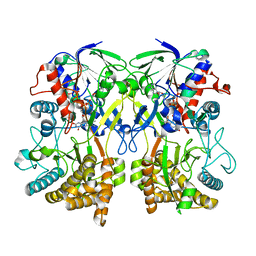 | | Knockout of GMC-oxidoreductase genes reveals that functional redundancy preserves mimivirus essential functions | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Alempic, J.M, Bisio, H, Villalta, A, Santini, S, Lartigue, A, Schmitt, A, Bugnot, C, Notaro, A, Belmudes, L, Adrait, A, Poirot, O, Ptchelkine, D, De Castro, C, Coute, Y, Abergel, C. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Functional redundancy revealed by the deletion of the mimivirus GMC-oxidoreductase genes.
Microlife, 5, 2024
|
|
8ORS
 
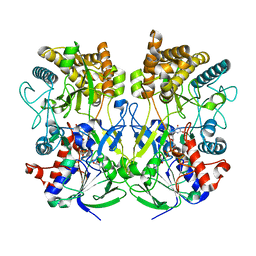 | | Knockout of GMC-oxidoreductase genes reveals that functional redundancy preserves mimivirus essential functions | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Alempic, J.M, Bisio, H, Villalta, A, Santini, S, Lartigue, A, Schmitt, A, Bugnot, C, Notaro, A, Belmudes, L, Adrait, A, Poirot, O, Ptchelkine, D, De Castro, C, Coute, Y, Abergel, C. | | Deposit date: | 2023-04-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Functional redundancy revealed by the deletion of the mimivirus GMC-oxidoreductase genes.
Microlife, 5, 2024
|
|
4U4I
 
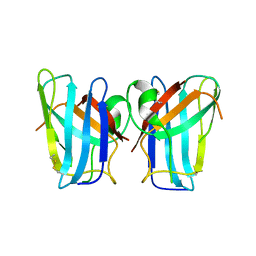 | | Megavirus chilensis superoxide dismutase | | Descriptor: | Cu/Zn superoxide dismutase | | Authors: | Lartigue, A, Claverie, J.-M, Burlat, B, Coutard, B, Abergel, C. | | Deposit date: | 2014-07-23 | | Release date: | 2014-11-05 | | Last modified: | 2014-12-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The megavirus chilensis cu,zn-superoxide dismutase: the first viral structure of a typical cellular copper chaperone-independent hyperstable dimeric enzyme.
J.Virol., 89, 2015
|
|
7YX3
 
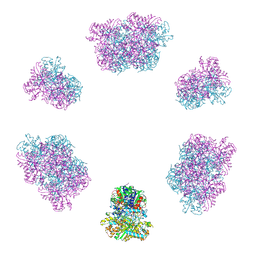 | | Structure of the Mimivirus genomic fibre in its compact 6-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30-nm diameter helical protein shield.
Elife, 11, 2022
|
|
7YX4
 
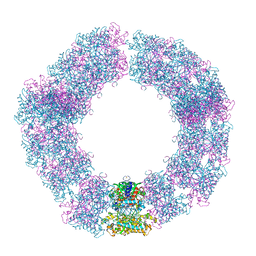 | | Structure of the Mimivirus genomic fibre in its compact 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
4XUL
 
 | | Crystal structure of M. chilensis Mg662 protein complexed with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Lartigue, A, Priet, S, Claverie, J.M, Abergel, C. | | Deposit date: | 2015-01-26 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of M. chilensis Mg662 protein complexed with GTP
To Be Published
|
|
4P37
 
 | | Crystal structure of the Megavirus polyadenylate synthase | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Priet, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2014-03-06 | | Release date: | 2015-04-01 | | Last modified: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | mRNA maturation in giant viruses: variation on a theme.
Nucleic Acids Res., 43, 2015
|
|
3KIP
 
 | | Crystal structure of type-II 3-dehydroquinase from C. albicans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-dehydroquinase, type II, ... | | Authors: | Trapani, S, Schoehn, G, Navaza, J, Abergel, C. | | Deposit date: | 2009-11-02 | | Release date: | 2010-05-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Macromolecular crystal data phased by negative-stained electron-microscopy reconstructions.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
