2OOH
 
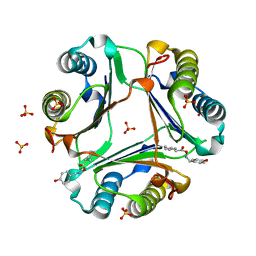 | | Crystal Structure of MIF bound to a Novel Inhibitor, OXIM-11 | | Descriptor: | 4-HYDROXYBENZALDEHYDE O-(CYCLOHEXYLCARBONYL)OXIME, GLYCEROL, Macrophage migration inhibitory factor, ... | | Authors: | Crichlow, G.V, Al-Abed, Y, Lolis, E. | | Deposit date: | 2007-01-25 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Alternative chemical modifications reverse the binding orientation of a pharmacophore scaffold in the active site of macrophage migration inhibitory factor.
J.Biol.Chem., 282, 2007
|
|
2DQA
 
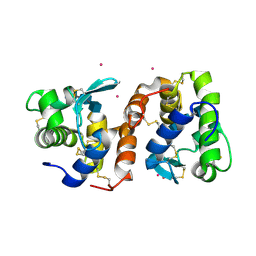 | | Crystal Structure of Tapes japonica Lysozyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme, PLATINUM (II) ION, ... | | Authors: | Goto, T, Kakuta, Y, Abe, Y, Takeshita, K, Imoto, T, Ueda, T. | | Deposit date: | 2006-05-24 | | Release date: | 2007-06-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Tapes japonica Lysozyme with Substrate Analogue: STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND MANIFESTATION OF ITS CHITINASE ACTIVITY ACCOMPANIED BY QUATERNARY STRUCTURAL CHANGE
J.Biol.Chem., 282, 2007
|
|
5XHL
 
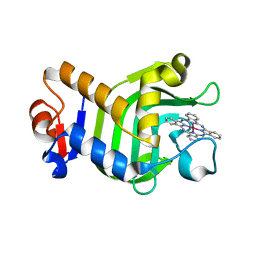 | | Crystal Structure of HasAp with Gallium Phthalocyanine | | Descriptor: | Heme acquisition protein HasAp, Phthalocyanine containing GA | | Authors: | Shoji, O, Shisaka, Y, Iwai, Y, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-04-21 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of HasAp with Gallium Phthalocyanine
to be published
|
|
7ARN
 
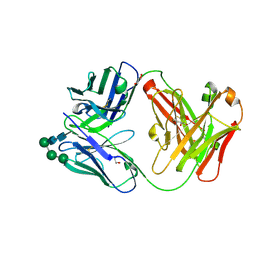 | | Crystal Structure of the Fab Fragment of a Glycosylated Lymphoma Antibody | | Descriptor: | Antibody Fab Fragment Heavy Chain, Antibody Fab Fragment Light Chain, GLYCEROL, ... | | Authors: | Pryce, R, Allen, J.D, Watanabe, Y, Crispin, M, Bowden, T.A. | | Deposit date: | 2020-10-25 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal Structure of the Fab Fragment of a Glycosylated Lymphoma Antibody
To Be Published
|
|
5XIE
 
 | | Crystal Structure of HasAp with 5-ethynyl-10,20-diphenylporphyrin | | Descriptor: | 5-Ethynyl-10,20-diphenylporphyrin containing FE, Heme acquisition protein HasAp | | Authors: | Shoji, O, Uehara, H, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-04-26 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of the Heme Acquisition Protein HasA with Iron(III)-5,15-Diphenylporphyrin and Derivatives Thereof as an Artificial Prosthetic Group
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4LUD
 
 | | Crystal Structure of HCK in complex with the fluorescent compound SKF86002 | | Descriptor: | 6-(4-fluorophenyl)-5-(pyridin-4-yl)-2,3-dihydroimidazo[2,1-b][1,3]thiazole, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-25 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4WCO
 
 | | Crystal structure of extracellular domain of human lectin-like transcript 1 (LLT1), the ligand for natural killer receptor-P1A | | Descriptor: | ACETATE ION, C-type lectin domain family 2 member D, SULFATE ION, ... | | Authors: | Kita, S, Matsubara, H, Kasai, Y, Tamaoki, T, Okabe, Y, Fukuhara, H, Kamishikiryo, J, Ose, T, Kuroki, K, Maenaka, K. | | Deposit date: | 2014-09-05 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structure of extracellular domain of human lectin-like transcript 1 (LLT1), the ligand for natural killer receptor-P1A
Eur.J.Immunol., 45, 2015
|
|
5XHJ
 
 | | Crystal Structure of P450BM3 with 5-Cyclohexylvaleroyl-L-Tryptophan | | Descriptor: | 5-cyclohexylpentanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Suzuki, K, Shoji, O, Stanfield, J.K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-04-21 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Control of stereoselectivity of benzylic hydroxylation catalysed by wild-type cytochrome P450BM3 using decoy molecules
CATALYSIS SCIENCE AND TECHNOLOGY, 7, 2017
|
|
5B2X
 
 | | Crystal Structure of P450BM3 mutant with N-perfluoroheptanoyl-L-tryptophan | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-2-[2,2,3,3,4,4,5,5,6,6,7,7,7-tridecakis(fluoranyl)heptanoylamino]propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | Authors: | Cong, Z, Shoji, O, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2016-02-03 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of P450BM3 with decoy molecules
to be published
|
|
5B2Y
 
 | | Crystal Structure of P450BM3 with N-perfluorodecanoyl-L-tryptophan | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-2-[2,2,3,3,4,4,5,5,6,6,7,7,8,8,9,9,10,10,10-nonadecakis(fluoranyl)decanoylamino]propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cong, Z, Shoji, O, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2016-02-07 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of P450BM3 with decoy molecules
to be published
|
|
5B2U
 
 | | Crystal Structure of P450BM3 with N-perfluorohexanoyl -L-tryptophan | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-2-[2,2,3,3,4,4,5,5,6,6,6-undecakis(fluoranyl)hexanoylamino]propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | Authors: | Cong, Z, Shoji, O, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2016-02-03 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of P450BM3 with decoy molecules
to be published
|
|
5AON
 
 | | Crystal structure of the conserved N-terminal domain of Pex14 from Trypanosoma brucei | | Descriptor: | PEROXIN 14, SULFATE ION | | Authors: | Obita, T, Sugawara, Y, Mizuguchi, M, Watanabe, Y, Kawaguchi, K, Imanaka, T. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.646 Å) | | Cite: | Characterization of the Interaction between Trypanosoma Brucei Pex5P and its Receptor Pex14P.
FEBS Lett., 590, 2016
|
|
5B2W
 
 | | Crystal Structure of P450BM3 with N-perfluorododecanoyl-L-tryptophan | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-2-[2,2,3,3,4,4,5,5,6,6,7,7,8,8,9,9,10,10,11,11,12,12,12-tricosakis(fluoranyl)dodecanoylamino]propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cong, Z, Shoji, O, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2016-02-03 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of P450BM3 with decoy molecules
to be published
|
|
7B81
 
 | |
4WR5
 
 | | Crystal Structure of GST Mutated with Halogenated Tyrosine (7cGST-1) | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, SULFATE ION | | Authors: | Akasaka, R, Kawazoe, M, Tomabechi, Y, Ohtake, K, Itagaki, T, Takemoto, C, Shirouzu, M, Yokoyama, S, Sakamoto, K. | | Deposit date: | 2014-10-23 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Protein stabilization utilizing a redefined codon
Sci Rep, 5, 2015
|
|
4WR4
 
 | | Crystal Structure of GST Mutated with Halogenated Tyrosine (7bGST-1) | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, SULFATE ION | | Authors: | Akasaka, R, Kawazoe, M, Tomabechi, Y, Ohtake, K, Itagaki, T, Takemoto, C, Shirouzu, M, Yokoyama, S, Sakamoto, K. | | Deposit date: | 2014-10-23 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protein stabilization utilizing a redefined codon
Sci Rep, 5, 2015
|
|
6ILI
 
 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 8-oxo-dGTP at pH 6.5 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2018-10-18 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
5JR8
 
 | | Disposal of Iron by a Mutant form of Siderocalin NGAL | | Descriptor: | GLYCEROL, Neutrophil gelatinase-associated lipocalin, PHOSPHATE ION | | Authors: | Rupert, P.B, Strong, R.K, Barasch, J, Hollman, M, Deng, R, Hod, E.A, Abergel, R, Allred, B, Xu, K, Darrah, S, Tekabe, Y, Perlstein, A, Bruck, E, Stauber, J, Corbin, K, Buchen, C, Slavkovich, V, Graziano, J, Spitalnik, S, Qiu, A. | | Deposit date: | 2016-05-05 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Disposal of iron by a mutant form of lipocalin 2.
Nat Commun, 7, 2016
|
|
8GSR
 
 | | Crystal structure of L-2,4-diketo-3-deoxyrhamnonate hydrolase from Sphingomonas sp. (apo-form) | | Descriptor: | L-2,4-diketo-3-deoxyrhamnonate hydrolase, MAGNESIUM ION | | Authors: | Fukuhara, S, Watanabe, Y, Watanabe, S, Nishiwaki, H. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of l-2,4-Diketo-3-deoxyrhamnonate Hydrolase Involved in the Nonphosphorylated l-Rhamnose Pathway from Bacteria.
Biochemistry, 62, 2023
|
|
8GST
 
 | | Crystal structure of L-2,4-diketo-3-deoxyrhamnonate hydrolase from Sphingomonas sp. (pyruvate bound-form) | | Descriptor: | L-2,4-diketo-3-deoxyrhamnonate hydrolase, MAGNESIUM ION, PYRUVIC ACID | | Authors: | Fukuhara, S, Watanabe, Y, Watanabe, S, Nishiwaki, H. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structure of l-2,4-Diketo-3-deoxyrhamnonate Hydrolase Involved in the Nonphosphorylated l-Rhamnose Pathway from Bacteria.
Biochemistry, 62, 2023
|
|
5AVM
 
 | | Crystal structures of 5-aminoimidazole ribonucleotide (AIR) synthetase, PurM, from Thermus thermophilus | | Descriptor: | Phosphoribosylformylglycinamidine cyclo-ligase, SULFATE ION | | Authors: | Kanagawa, M, Baba, S, Watanabe, Y, Nakagawa, N, Ebihara, A, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2015-06-23 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and ligand binding of PurM proteins from Thermus thermophilus and Geobacillus kaustophilus
J.Biochem., 159, 2016
|
|
6IN0
 
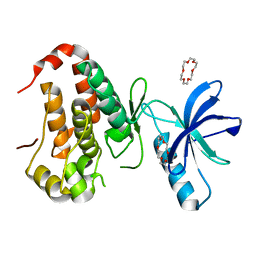 | | Crystal structure of EphA3 in complex with 18-Crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, CHLORIDE ION, Ephrin type-A receptor 3 | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitor
To Be Published
|
|
6IN4
 
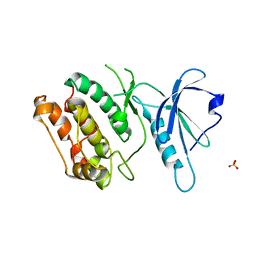 | | Crystal structure of apo DAPK1 in the presence of 18-crown-6 | | Descriptor: | Death-associated protein kinase 1, SULFATE ION | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitor
To Be Published
|
|
6IMZ
 
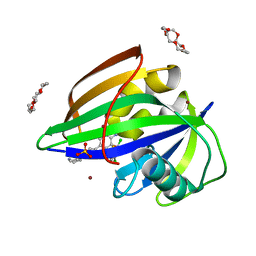 | | Crystal structure of MTH1 in complex with 18-Crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase, ... | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitor
To Be Published
|
|
6IN3
 
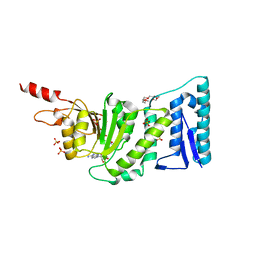 | | Crystal structure of DOT1L in complex with 18-Crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitor
To Be Published
|
|
