4CH5
 
 | | Structure of pyrrolysyl-tRNA synthetase in complex with adenylated propionyl lysine | | Descriptor: | (S)-2-amino-6-propionamidohexanoic(((2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl phosphoric) anhydride, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Fluegel, V, Vrabel, M, Schneider, S. | | Deposit date: | 2013-11-28 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Site-Specific Incorporation of Lysine Derivatives Into Proteins.
Plos One, 9, 2014
|
|
4CH3
 
 | | Structure of pyrrolysyl-tRNA synthetase in complex with adenylated butyryl lysine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-({[(2R)-2-amino-6-(butanoylamino)hexanoyl]oxy}phosphinato)adenosine, MAGNESIUM ION, ... | | Authors: | Fluegel, V, Vrabel, M, Schneider, S. | | Deposit date: | 2013-11-28 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Basis for the Site-Specific Incorporation of Lysine Derivatives Into Proteins.
Plos One, 9, 2014
|
|
1OSB
 
 | | Conjugative Relaxase TrwC in complex with OriT Dna. Metal-free structure. | | Descriptor: | Dna oligonucleotide, SULFATE ION, TrwC protein | | Authors: | Guasch, A, Lucas, M, Moncalian, G, Cabezas, M, Perez-Luque, R, Gomis-Ruth, F.X, de la Cruz, F, Coll, M. | | Deposit date: | 2003-03-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC.
Nat.Struct.Biol., 10, 2003
|
|
2HZ9
 
 | | Crystal structure of Lys12Val/Asn95Val/Cys117Val mutant of human acidic fibroblast growth factor at 1.70 angstrom resolution. | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Dubey, V.K, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2006-08-08 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Spackling the Crack: Stabilizing Human Fibroblast Growth Factor-1 by Targeting the N and C terminus beta-Strand Interactions
J.Mol.Biol., 371, 2007
|
|
4YOL
 
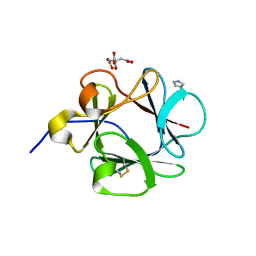 | | Human fibroblast growth factor-1 C16S/A66C/C117A/P134A | | Descriptor: | CITRATE ANION, Fibroblast growth factor 1, IMIDAZOLE | | Authors: | Xia, X, Blaber, M. | | Deposit date: | 2015-03-11 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Engineering a Cysteine-Free Form of Human Fibroblast Growth Factor-1 for "Second Generation" Therapeutic Application.
J.Pharm.Sci., 105, 2016
|
|
5AHV
 
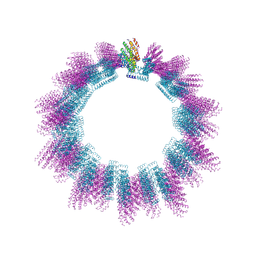 | | Cryo-EM structure of helical ANTH and ENTH tubules on PI(4,5)P2-containing membranes | | Descriptor: | ANTH DOMAIN OF ENDOCYTIC ADAPTOR SLA2, ENTH DOMAIN OF EPSIN ENT1 | | Authors: | Skruzny, M, Desfosses, A, Prinz, S, Dodonova, S.O, Gieras, A, Uetrecht, C, Jakobi, A.J, Abella, M, Hagen, W.J.H, Schulz, J, Meijers, R, Rybin, V, Briggs, J.A.G, Sachse, C, Kaksonen, M. | | Deposit date: | 2015-02-10 | | Release date: | 2015-05-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | An Organized Co-Assembly of Clathrin Adaptors is Essential for Endocytosis.
Dev.Cell, 33, 2015
|
|
2N7H
 
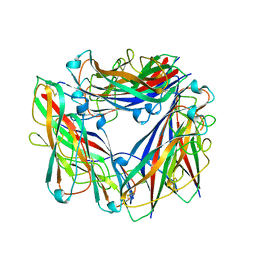 | | Hybrid structure of the Type 1 Pilus of Uropathogenic E.coli | | Descriptor: | FimA | | Authors: | Habenstein, B, Loquet, A, Giller, K, Vasa, S, Becker, S, Habeck, M, Lange, A. | | Deposit date: | 2015-09-11 | | Release date: | 2015-09-23 | | Last modified: | 2015-10-14 | | Method: | SOLID-STATE NMR | | Cite: | Hybrid Structure of the Type 1 Pilus of Uropathogenic Escherichia coli.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4CH6
 
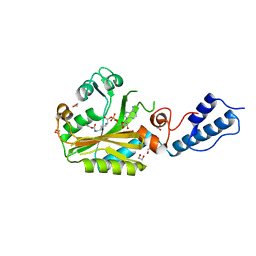 | | Structure of pyrrolysyl-tRNA synthetase in complex with adenylated propargyloxycarbonyl lysine | | Descriptor: | (S)-2-amino-6-(((prop-2-yn-1-yloxy)carbonyl)amino)hexanoic (((2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl phosphoric)anhydride, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Fluegel, V, Vrabel, M, Schneider, S. | | Deposit date: | 2013-11-28 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for the Site-Specific Incorporation of Lysine Derivatives Into Proteins.
Plos One, 9, 2014
|
|
4AB7
 
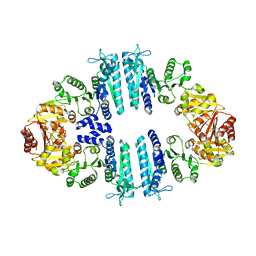 | | Crystal structure of a tetrameric acetylglutamate kinase from Saccharomyces cerevisiae complexed with its substrate N- acetylglutamate | | Descriptor: | N-ACETYL-L-GLUTAMATE, PROTEIN ARG5,6, MITOCHONDRIAL | | Authors: | de Cima, S, Gil-Ortiz, F, Crabeel, M, Fita, I, Rubio, V. | | Deposit date: | 2011-12-07 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Insight on an Arginine Synthesis Metabolon from the Tetrameric Structure of Yeast Acetylglutamate Kinase
Plos One, 7, 2012
|
|
1YBJ
 
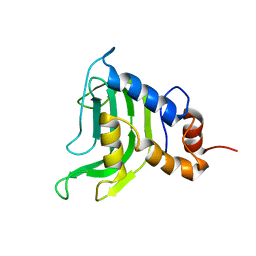 | | Structural and Dynamics studies of both apo and holo forms of the hemophore HasA | | Descriptor: | Hemophore HasA | | Authors: | Wolff, N, Izadi-Pruneyre, N, Couprie, J, Habeck, M, Linge, J, Rieping, W, Wandersman, C, Nilges, M, Delepierre, M, Lecroisey, A. | | Deposit date: | 2004-12-21 | | Release date: | 2005-12-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Comparative analysis of structural and dynamic properties of the loaded and unloaded hemophore HasA: functional implications.
J.Mol.Biol., 376, 2008
|
|
4QKR
 
 | |
4Q91
 
 | |
4CH4
 
 | | Structure of pyrrolysyl-tRNA synthetase in complex with adenylated crotonyl lysine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[({(2R)-2-amino-6-[(2E)-but-2-enoylamino]hexanoyl}oxy)phosphinato]adenosine, MAGNESIUM ION, ... | | Authors: | Fluegel, V, Vrabel, M, Schneider, S. | | Deposit date: | 2013-11-28 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Basis for the Site-Specific Incorporation of Lysine Derivatives Into Proteins.
Plos One, 9, 2014
|
|
2HWA
 
 | | Crystal structure of Lys12Thr/Cys117Val mutant of human acidic fibroblast growth factor at 1.65 angstrom resolution. | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Dubey, V.K, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2006-08-01 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Spackling the crack: stabilizing human fibroblast growth factor-1 by targeting the N and C terminus beta-strand interactions.
J.Mol.Biol., 371, 2007
|
|
2HW9
 
 | | Crystal structure of Lys12Cys/Cys117Val mutant of human acidic fibroblast Growth factor at 1.60 angstrom resolution. | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Dubey, V.K, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2006-08-01 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Spackling the crack: stabilizing human fibroblast growth factor-1 by targeting the N and C terminus beta-strand interactions.
J.Mol.Biol., 371, 2007
|
|
2HWM
 
 | | Crystal structure of Lys12Val/Cys117Val mutant of human acidic fibroblast growth factor at 1.60 angstrom resolution | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1 | | Authors: | Dubey, V.K, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2006-08-01 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Spackling the crack: stabilizing human fibroblast growth factor-1 by targeting the N and C terminus beta-strand interactions.
J.Mol.Biol., 371, 2007
|
|
4QKS
 
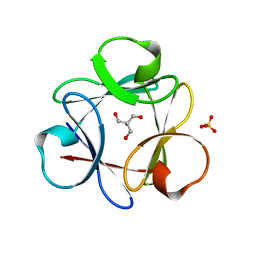 | | Crystal Structure of 6xTrp/PV2: de novo designed beta-trefoil architecture with symmetric primary structure (L22W/L44W/L64W/L85W/L108W/L132W his Primitive Version 2) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DE NOVO PROTEIN 6XTRP/PV2, SULFATE ION | | Authors: | Longo, L.M, Tenorio, C.A, Blaber, M. | | Deposit date: | 2014-06-09 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A single aromatic core mutation converts a designed "primitive" protein from halophile to mesophile folding.
Protein Sci., 24, 2015
|
|
1KWF
 
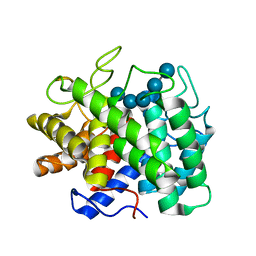 | | Atomic Resolution Structure of an Inverting Glycosidase in Complex with Substrate | | Descriptor: | Endoglucanase A, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Guerin, D.M.A, Lascombe, M.-B, Costabel, M, Souchon, H, Lamzin, V, Beguin, P, Alzari, P.M. | | Deposit date: | 2002-01-29 | | Release date: | 2002-03-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Atomic (0.94 A) resolution structure of an inverting glycosidase in complex with substrate.
J.Mol.Biol., 316, 2002
|
|
3C7C
 
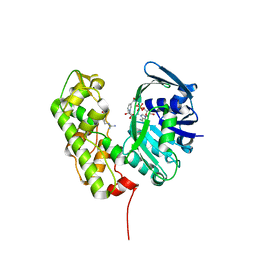 | | A structural basis for substrate and stereo selectivity in octopine dehydrogenase (ODH-NADH-L-Arginine) | | Descriptor: | ARGININE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Octopine dehydrogenase | | Authors: | Smits, S.H.J, Mueller, A, Schmitt, L, Grieshaber, M.K. | | Deposit date: | 2008-02-07 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A structural basis for substrate selectivity and stereoselectivity in octopine dehydrogenase from Pecten maximus.
J.Mol.Biol., 381, 2008
|
|
1M16
 
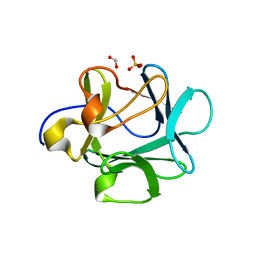 | | Human Acidic Fibroblast Growth Factor. 141 Amino Acid Form with Amino Terminal His Tag and Leu 44 Replaced with Phe (L44F), Leu 73 Replaced with Val (L73V), Val 109 Replaced with Leu (V109L) and Cys 117 Replaced with Val (C117V). | | Descriptor: | FORMIC ACID, SULFATE ION, acidic fibroblast growth factor | | Authors: | Brych, S.R, Kim, J, Spielmann, G.L, Logan, T.M, Blaber, M. | | Deposit date: | 2002-06-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Accommodation of a highly symmetric core within a symmetric protein superfold
Protein Sci., 12, 2003
|
|
3CRG
 
 | | Crystal structure of human fibroblast growth factor-1 with mutations Glu81Ala, Glu82Asn and Lys101Ala | | Descriptor: | FORMIC ACID, GLYCEROL, Heparin-binding growth factor 1, ... | | Authors: | Meher, A.K, Honjo, E, Kuroki, R, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2008-04-07 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering an improved crystal contact across a solvent-mediated interface of human fibroblast growth factor 1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1IQ8
 
 | | Crystal Structure of archaeosine tRNA-guanine transglycosylase from Pyrococcus horikoshii | | Descriptor: | ARCHAEOSINE TRNA-GUANINE TRANSGLYCOSYLASE, MAGNESIUM ION, ZINC ION | | Authors: | Ishitani, R, Nureki, O, Fukai, S, Kijimoto, T, Nameki, N, Watanabe, M, Kondo, H, Sekine, M, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-07-09 | | Release date: | 2002-05-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of archaeosine tRNA-guanine transglycosylase.
J.Mol.Biol., 318, 2002
|
|
1YTO
 
 | | Crystal Structure of Gly19 deletion Mutant of Human Acidic Fibroblast Growth Factor | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Lee, J, Blaber, M. | | Deposit date: | 2005-02-10 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conversion of type I 4:6 to 3:5 beta-turn types in human acidic fibroblast growth factor: Effects upon structure, stability, folding, and mitogenic function.
Proteins, 62, 2006
|
|
1UAW
 
 | | Solution structure of the N-terminal RNA-binding domain of mouse Musashi1 | | Descriptor: | mouse-musashi-1 | | Authors: | Miyanoiri, Y, Kobayashi, H, Watanabe, M, Ikeda, T, Nagata, T, Okano, H, Uesugi, S, Katahira, M. | | Deposit date: | 2003-03-24 | | Release date: | 2004-03-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Origin of higher affinity to RNA of the N-terminal RNA-binding domain than that of the C-terminal one of a mouse neural protein, musashi1, as revealed by comparison of their structures, modes of interaction, surface electrostatic potentials, and backbone dynamics
J.Biol.Chem., 278, 2003
|
|
1S6M
 
 | | Conjugative Relaxase Trwc In Complex With Orit DNA. Metal-Bound Structure | | Descriptor: | DNA (25-MER), NICKEL (II) ION, TrwC | | Authors: | Guasch, A, Lucas, M, Moncalian, G, Cabezas, M, Perez-Luque, R, Gomis-Ruth, F.X, de la Cruz, F, Coll, M. | | Deposit date: | 2004-01-26 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Unveiling the molecular mechanism of a conjugative relaxase: The structure of TrwC complexed with a 27-mer DNA comprising the recognition hairpin and the cleavage site.
J.Mol.Biol., 358, 2006
|
|
