8GCR
 
 | | HPV16 E6-E6AP-p53 complex | | Descriptor: | Cellular tumor antigen p53, Maltose/maltodextrin-binding periplasmic protein,Protein E6, Ubiquitin-protein ligase E3A, ... | | Authors: | Bratkowski, M.A, Wang, J.C.K, Hao, Q, Nile, A.H. | | Deposit date: | 2023-03-02 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structure of the p53 degradation complex from HPV16.
Nat Commun, 15, 2024
|
|
5HXN
 
 | | Crystal Structure of Z,Z-Farnesyl Diphosphate Synthase (D71M and E75A mutants) from the Wild Tomato Solanum habrochaites | | Descriptor: | (2Z,6Z)-farnesyl diphosphate synthase, chloroplastic | | Authors: | Lee, C.C, Chan, Y.T, Wang, A.H.J. | | Deposit date: | 2016-01-31 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure and Potential Head-to-Middle Condensation Function of aZ,Z-Farnesyl Diphosphate Synthase.
Acs Omega, 2, 2017
|
|
8I4F
 
 | | Omicron spike variant XBB with n3130v-Fc | | Descriptor: | Spike glycoprotein, n3130v-Fc | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
5HXO
 
 | | Crystal Structure of Z,Z-Farnesyl Diphosphate Synthase with D71M, E75A and H103Y Mutants | | Descriptor: | (2Z,6Z)-farnesyl diphosphate synthase, chloroplastic | | Authors: | Lee, C.C, Chan, Y.T, Wang, A.H.J. | | Deposit date: | 2016-01-31 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure and Potential Head-to-Middle Condensation Function of aZ,Z-Farnesyl Diphosphate Synthase.
Acs Omega, 2, 2017
|
|
8I4G
 
 | | Omicron spike variant BQ.1.1 with n3130v-Fc | | Descriptor: | Spike glycoprotein, n3130v-Fc | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
8I4E
 
 | | Omicron spike variant XBB with Bn03 | | Descriptor: | Bn03, Spike glycoprotein | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
5HXT
 
 | | Crystal Structure of Z,Z-Farnesyl Diphosphate Synthase (D71M, E75A and H103Y Mutants) Complexed with IPP and DMSPP | | Descriptor: | (2Z,6Z)-farnesyl diphosphate synthase, chloroplastic, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, ... | | Authors: | Lee, C.C, Chan, Y.T, Wang, A.H.J. | | Deposit date: | 2016-01-31 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure and Potential Head-to-Middle Condensation Function of a Z,Z-Farnesyl Diphosphate Synthase
Acs Omega, 2, 2017
|
|
8I4H
 
 | | Omicron spike variant BA.1 with Bn03 | | Descriptor: | Bn03, Spike glycoprotein | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
5HXQ
 
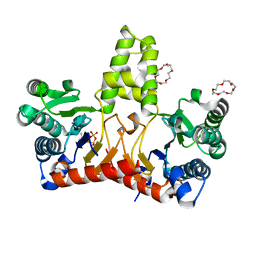 | | Crystal Structure of Z,Z-Farnesyl Diphosphate Synthase (D71M, E75A and H103Y Mutants) Complexed with DMSPP | | Descriptor: | (2Z,6Z)-farnesyl diphosphate synthase, chloroplastic, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, ... | | Authors: | Lee, C.C, Chan, Y.T, Wang, A.H.J. | | Deposit date: | 2016-01-31 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure and Potential Head-to-Middle Condensation Function of a Z,Z-Farnesyl Diphosphate Synthase
Acs Omega, 2, 2017
|
|
5G50
 
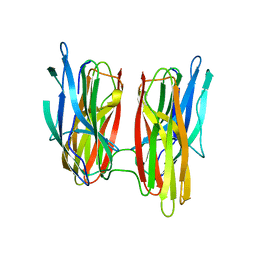 | |
5FD3
 
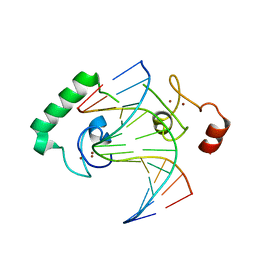 | | Structure of Lin54 tesmin domain bound to DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*TP*TP*CP*AP*AP*AP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*TP*TP*TP*GP*AP*AP*AP*CP*T)-3'), Protein lin-54 homolog, ... | | Authors: | Marceau, A.H, Felthousen, J.G, Goetsch, P.D, Lee, H, Tripathi, S.M, Strome, S, Litovchick, L, Rubin, S.M. | | Deposit date: | 2015-12-15 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis for LIN54 recognition of CHR elements in cell cycle-regulated promoters.
Nat Commun, 7, 2016
|
|
5HXP
 
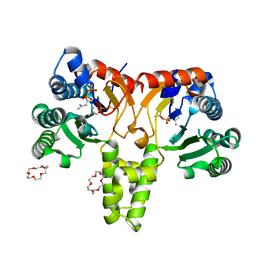 | | Crystal Structure of Z,Z-Farnesyl Diphosphate Synthase (D71M, E75A and H103Y Mutants) Complexed with IPP | | Descriptor: | (2Z,6Z)-farnesyl diphosphate synthase, chloroplastic, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, ... | | Authors: | Lee, C.C, Chan, Y.T, Wang, A.H.J. | | Deposit date: | 2016-01-31 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure and Potential Head-to-Middle Condensation Function of aZ,Z-Farnesyl Diphosphate Synthase.
Acs Omega, 2, 2017
|
|
5G64
 
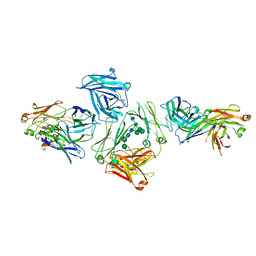 | | The complex between human IgE-Fc and two anti-IgE Fab fragments | | Descriptor: | FAB FRAGMENT, IG EPSILON CHAIN C REGION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Davies, A.M, Allan, E.G, Keeble, A.H, Delgado, J, Cossins, B.P, Mitropoulou, A.N, Pang, M.O.Y, Ceska, T, Beavil, A.J, Craggs, G, Westwood, M, Henry, A.J, McDonnell, J.M, Sutton, B.J. | | Deposit date: | 2016-06-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.715 Å) | | Cite: | Allosteric mechanism of action of the therapeutic anti-IgE antibody omalizumab.
J. Biol. Chem., 292, 2017
|
|
5G2N
 
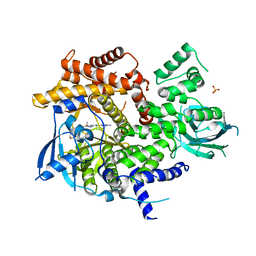 | | X-ray structure of PI3Kinase Gamma in complex with Copanlisib | | Descriptor: | 2-azanyl-~{N}-[7-methoxy-8-(3-morpholin-4-ylpropoxy)-2,3-dihydroimidazo[1,2-c]quinazolin-5-yl]pyrimidine-5-carboxamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM, SULFATE ION | | Authors: | Schaefer, M, Scott, W.J, Hentemann, M.F, Rowley, R.B, Bull, C.O, Jenkins, S, Bullion, A.M, Johnson, J, Redman, A, Robbins, A.H, Esler, W, Fracasso, R.P, Garrison, T, Hamilton, M, Michels, M, Wood, J.E, Wilkie, D.P, Xiao, H, Levy, J, Liu, N, Stasik, E, Brands, M, Lefranc, J. | | Deposit date: | 2016-04-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery and Sar of Novel 2,3-Dihydroimidazo(1,2-C)Quinazoline Pi3K Inhibitors: Identification of Copanlisib (Bay 80-6946)
Chemmedchem, 11, 2016
|
|
6US6
 
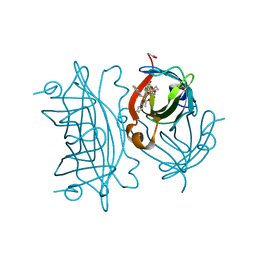 | | Artificial Iron Proteins: Modelling the Active Sites in Non-Heme Dioxygenases | | Descriptor: | ACETATE ION, Streptavidin, {N-(2-{bis[(pyridin-2-yl-kappaN)methyl]amino-kappaN}ethyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}iron(3+) | | Authors: | Miller, K.R, Paretsky, J.D, Follmer, A.H, Heinisch, T, Mittra, K, Gul, S, Kim, I.-S, Fuller, F.D, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bhowmick, A, Sauter, N.K, Kern, J, Yano, J, Green, M.T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2019-10-24 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Artificial Iron Proteins: Modeling the Active Sites in Non-Heme Dioxygenases.
Inorg.Chem., 59, 2020
|
|
6VP1
 
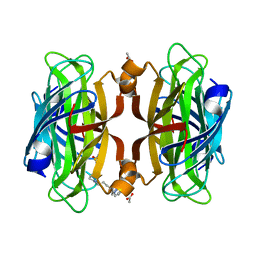 | | Artificial Metalloproteins with Dinuclear Iron Centers | | Descriptor: | ACETATE ION, Streptavidin, {N-(4-{bis[(pyridin-2-yl-kappaN)methyl]amino-kappaN}butyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}iron(3+) | | Authors: | Miller, K.R, Follmer, A.H, Jasniewski, A.J, Sabuncu, S, Biswas, S, Albert, T, Hendrich, M.P, Moenne-Loccoz, P, Borovik, A.S. | | Deposit date: | 2020-02-01 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Artificial Metalloproteins with Dinuclear Iron-Hydroxido Centers.
J.Am.Chem.Soc., 143, 2021
|
|
6VP2
 
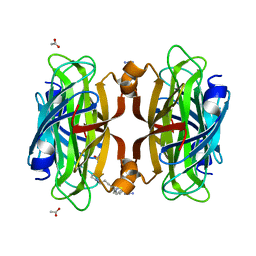 | | Artificial Metalloproteins with Dinuclear Iron Centers | | Descriptor: | ACETATE ION, AZIDE ION, Streptavidin, ... | | Authors: | Miller, K.R, Follmer, A.H, Jasniewski, A.J, Sabuncu, S, Biswas, S, Albert, T, Hendrich, M.P, Moenne-Loccoz, P, Borovik, A.S. | | Deposit date: | 2020-02-01 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Artificial Metalloproteins with Dinuclear Iron-Hydroxido Centers.
J.Am.Chem.Soc., 143, 2021
|
|
6VO9
 
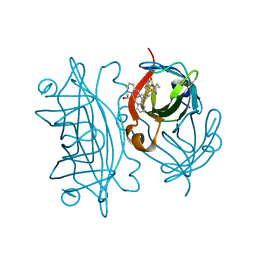 | | Artificial Metalloproteins with Dinuclear Iron Centers | | Descriptor: | ACETATE ION, Streptavidin, {N-(4-{bis[(pyridin-2-yl-kappaN)methyl]amino-kappaN}butyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}iron(3+) | | Authors: | Miller, K.R, Follmer, A.H, Jasniewski, A.J, Sabuncu, S, Biswas, S, Albert, T, Hendrich, M.P, Moenne-Loccoz, P, Borovik, A.S. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Artificial Metalloproteins with Dinuclear Iron-Hydroxido Centers.
J.Am.Chem.Soc., 143, 2021
|
|
6W25
 
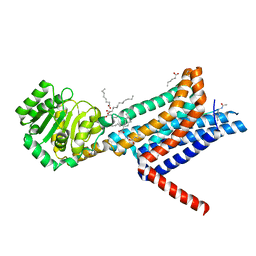 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with SHU9119 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4,GlgA glycogen synthase,Melanocortin receptor 4, OLEIC ACID, ... | | Authors: | Yu, J, Gimenez, L.E, Hernandez, C.C, Wu, Y, Wein, A.H, Han, G.W, McClary, K, Mittal, S.R, Burdsall, K, Stauch, B, Wu, L, Stevens, S.N, Peisley, A, Williams, S.Y, Chen, V, Millhauser, G.L, Zhao, S, Cone, R.D, Stevens, R.C. | | Deposit date: | 2020-03-04 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Determination of the melanocortin-4 receptor structure identifies Ca2+as a cofactor for ligand binding.
Science, 368, 2020
|
|
6VOB
 
 | | Artificial Metalloproteins with Dinuclear Iron Centers | | Descriptor: | ACETATE ION, AZIDE ION, Streptavidin, ... | | Authors: | Miller, K.R, Follmer, A.H, Jasniewski, A.J, Sabuncu, S, Biswas, S, Albert, T, Hendrich, M.P, Moenne-Loccoz, P, Borovik, A.S. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Artificial Metalloproteins with Dinuclear Iron-Hydroxido Centers.
J.Am.Chem.Soc., 143, 2021
|
|
6X4C
 
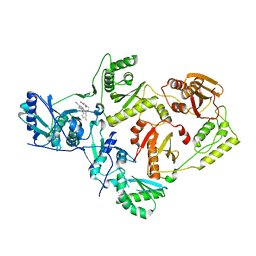 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-fluorophenoxy)-5,8-dimethyl-2-naphthonitrile (JLJ658), a Non-nucleoside Inhibitor | | Descriptor: | 7-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-5,8-dimethylnaphthalene-2-carbonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.861 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
6X4D
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(cyclopropylmethyl)-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile (JLJ678), a Non-nucleoside Inhibitor | | Descriptor: | 5-(cyclopropylmethyl)-7-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-8-methylnaphthalene-2-carbonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
6X4A
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with 5-chloro-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile (JLJ651), a Non-nucleoside Inhibitor | | Descriptor: | 5-chloro-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.537 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
6X49
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with 7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile (JLJ649), a Non-nucleoside Inhibitor | | Descriptor: | 7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.745 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
6X4F
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with methyl 2-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)acetate (JLJ681), a Non-nucleoside Inhibitor | | Descriptor: | Reverse transcriptase/ribonuclease H, SULFATE ION, methyl (6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)acetate, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
