1LUM
 
 | | NMR Structure of the Itk SH2 domain, Pro287trans, 20 low energy structures | | Descriptor: | Tyrosine-protein kinase ITK/TSK | | Authors: | Mallis, R.J, Brazin, K.N, Fulton, D.B, Andreotti, A.H. | | Deposit date: | 2002-05-22 | | Release date: | 2002-11-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of a proline-driven conformational switch
within the Itk SH2 domain
Nat.Struct.Biol., 9, 2002
|
|
2LSH
 
 | |
8F4A
 
 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with chlorhexidine | | Descriptor: | 1-[6-[azanylidene-[[azanylidene-[[(4-chlorophenyl)amino]methyl]-$l^{4}-azanyl]methyl]-$l^{4}-azanyl]hexyl]-3-[~{N}-(4-chlorophenyl)carbamimidoyl]guanidine, GLYCEROL, N-acetyltransferase Eis | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-10 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
8F55
 
 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis H37Rv in complex with inhibitor SGT1614 | | Descriptor: | (2E)-2-[(4-chlorophenyl)methylidene]hydrazine-1-carboximidamide, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
8F57
 
 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with inhibitor SGT1615 | | Descriptor: | (1E)-1-[(4-chlorophenyl)methylidene]-2-(3-fluorophenyl)hydrazine, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-12 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
3HS4
 
 | | Human carbonic anhydrase II complexed with acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Robbins, A.H, Genis, C, Domsic, J, Sippel, K.H, Agbandje-McKenna, M, McKenna, R. | | Deposit date: | 2009-06-10 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-resolution structure of human carbonic anhydrase II complexed with acetazolamide reveals insights into inhibitor drug design.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
8F4W
 
 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with venlafaxine | | Descriptor: | 1-[(1R)-2-(dimethylamino)-1-(4-methoxyphenyl)ethyl]cyclohexan-1-ol, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
8F4T
 
 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with proguanil | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Punetha, A, Pang, A.H, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
2L75
 
 | |
8F58
 
 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with inhibitor SGT1616 | | Descriptor: | (1E)-1-[(4-chlorophenyl)methylidene]-2-(4-fluorophenyl)hydrazine, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-12 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
8F4Z
 
 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with chloroquine | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N-acetyltransferase Eis, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
8F4U
 
 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with azelastine | | Descriptor: | 4-[(4-chlorophenyl)methyl]-2-[(4S)-1-methylazepan-4-yl]phthalazin-1(2H)-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
8F51
 
 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with mefloquine | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
8EUH
 
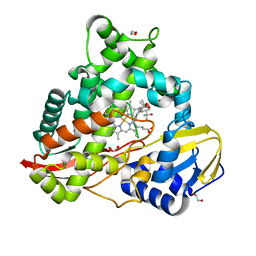 | | cytochrome P450terp (cyp108A1) bound to alpha-terpineol | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450-terp, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gable, J.A, Follmer, A.H, Poulos, T.L. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cooperative Substrate Binding Controls Catalysis in Bacterial Cytochrome P450terp (CYP108A1).
J.Am.Chem.Soc., 2023
|
|
8EUK
 
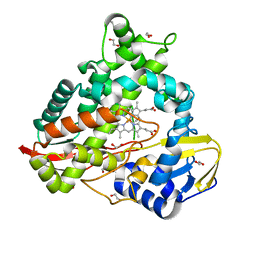 | |
8EUL
 
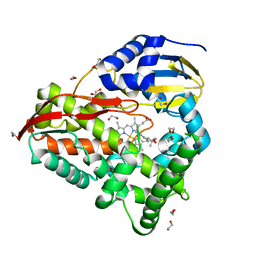 | | cytochrome P450terp (cyp108A1) mutant F188A bound to alpha-terpineol | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450-terp, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gable, J.A, Follmer, A.H, Poulos, T.L. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Cooperative Substrate Binding Controls Catalysis in Bacterial Cytochrome P450terp (CYP108A1).
J.Am.Chem.Soc., 2023
|
|
3HZD
 
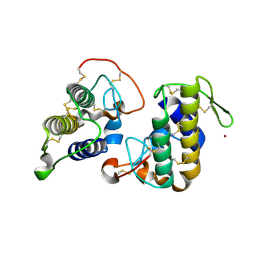 | | Crystal structure of bothropstoxin-I (BthTX-I), a PLA2 homologue from Bothrops jararacussu venom | | Descriptor: | LITHIUM ION, Phospholipase A2 homolog bothropstoxin-1 | | Authors: | Silva, M.C.O, Marchi-Salvador, D.P, Fernandes, C.A.H, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
2M0X
 
 | |
8EIZ
 
 | | Cryo-EM structure of squid sensory receptor CRB1 | | Descriptor: | N-benzyl-2-(2,6-dimethylanilino)-N,N-diethyl-2-oxoethan-1-aminium, Squid sensory receptor CRB1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kang, G, Kim, J.J, Allard, C.A.H, Valencia-Montoya, W.A, van Giesen, L, Kilian, P.B, Bai, X, Bellono, N.W, Hibbs, R.E. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Sensory specializations drive octopus and squid behaviour.
Nature, 616, 2023
|
|
8EIS
 
 | | Cryo-EM structure of octopus sensory receptor CRT1 | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Octopus sensory receptor | | Authors: | Kang, G, Kim, J.J, Allard, C.A.H, Valencia-Montoya, W.A, Bellono, N.W, Hibbs, R.E. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Sensory specializations drive octopus and squid behaviour.
Nature, 616, 2023
|
|
3HSQ
 
 | |
3I3X
 
 | |
3HZW
 
 | | Crystal structure of bothropstoxin-I chemically modified by p-bromophenacyl bromide (BPB) | | Descriptor: | ISOPROPYL ALCOHOL, Phospholipase A2 homolog bothropstoxin-1, p-Bromophenacyl bromide | | Authors: | Fernandes, C.A.H, Marchi-Salvador, D.P, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
8EPW
 
 | | Crystal Structure of KRAS4b-G13D (GMPPNP-bound) in complex with RAS-binding domain (RBD) of RAF1/CRAF | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Tran, T.H, Chan, A.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2022-10-06 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reduced dynamic complexity allows structure elucidation of an excited state of KRAS G13D .
Commun Biol, 6, 2023
|
|
2MBV
 
 | | LMO4-LIM2 in complex with DEAF1 (404-418) | | Descriptor: | Fusion protein of LIM domain transcription factor LMO4 (77-147) and DEAF1 (404-418), ZINC ION | | Authors: | Joseph, S, Matthews, J.M, Kwan, A.H.-Y, Mackay, J.P, Cubeddu, L, Foo, P. | | Deposit date: | 2013-08-06 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural characterisation of LIM-only protein 4 (LMO4) in complex with Deformed Epidermal Autoregulatory Factor-1 (DEAF1)
To be Published
|
|
