2C3V
 
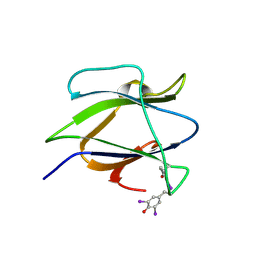 | | Structure of iodinated CBM25 from Bacillus halodurans amylase | | Descriptor: | ALPHA-AMYLASE G-6, IODIDE ION | | Authors: | Boraston, A.B, Healey, M, Klassen, J, Ficko-Blean, E, Lammerts van Bueren, A, Law, V. | | Deposit date: | 2005-10-12 | | Release date: | 2005-10-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A Structural and Functional Analysis of Alpha-Glucan Recognition by Family 25 and 26 Carbohydrate-Binding Modules Reveals a Conserved Mode of Starch Recognition
J.Biol.Chem., 281, 2006
|
|
2BW7
 
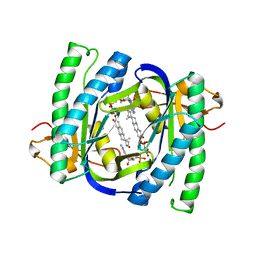 | | A novel mechanism for adenylyl cyclase inhibition from the crystal structure of its complex with catechol estrogen | | Descriptor: | 2,3,17BETA-TRIHYDROXY-1,3,5(10)-ESTRATRIENE, ADENYLATE CYCLASE, CALCIUM ION, ... | | Authors: | Steegborn, C, Litvin, T.N, Hess, K.C, Capper, A.B, Taussig, R, Buck, J, Levin, L.R, Wu, H. | | Deposit date: | 2005-07-12 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Novel Mechanism for Adenylyl Cyclase Inhibition from the Crystal Structure of its Complex with Catechol Estrogen
J.Biol.Chem., 280, 2005
|
|
2C3X
 
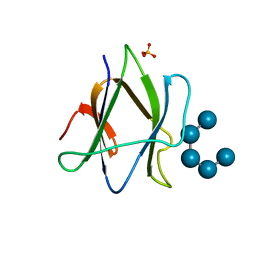 | | Structure of iodinated CBM25 from Bacillus halodurans amylase in complex with maltotetraose | | Descriptor: | ALPHA-AMYLASE G-6, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Boraston, A.B, Healey, M, Klassen, J, Ficko-Blean, E, Lammerts van Bueren, A, Law, V. | | Deposit date: | 2005-10-12 | | Release date: | 2005-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Structural and Functional Analysis of Alpha-Glucan Recognition by Family 25 and 26 Carbohydrate-Binding Modules Reveals a Conserved Mode of Starch Recognition
J.Biol.Chem., 281, 2006
|
|
7QWV
 
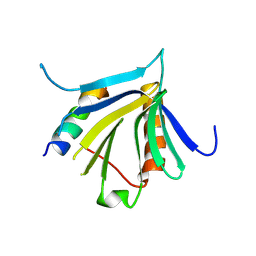 | | Crystal structure of the REC114-TOPOVIBL complex. | | Descriptor: | Meiotic recombination protein REC114, Type 2 DNA topoisomerase 6 subunit B-like | | Authors: | Juarez-Martinez, A.B, Robert, T, de Massy, B, Kadlec, J. | | Deposit date: | 2022-01-25 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | TOPOVIBL-REC114 interaction regulates meiotic DNA double-strand breaks.
Nat Commun, 13, 2022
|
|
7RSO
 
 | | AMC016 SOSIP.v4.2 in complex with PGV04 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMC016 gp120, ... | | Authors: | Bader, D.L.V, Cottrell, C.A, Ward, A.B. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-15 | | Last modified: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The Glycan Hole Area of HIV-1 Envelope Trimers Contributes Prominently to the Induction of Autologous Neutralization.
J.Virol., 96, 2022
|
|
7RSN
 
 | | AMC018 SOSIP.v4.2 in complex with PGV04 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMC018 gp120, ... | | Authors: | Cottrell, C.A, Ward, A.B. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-15 | | Last modified: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The Glycan Hole Area of HIV-1 Envelope Trimers Contributes Prominently to the Induction of Autologous Neutralization.
J.Virol., 96, 2022
|
|
2BR7
 
 | | Crystal Structure of Acetylcholine-binding Protein (AChBP) from Aplysia californica in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Celie, P.H.N, Kasheverov, I.E, Mordvintsev, D.Y, Hogg, R.C, Van Nierop, P, Van Elk, R, Van Rossum-Fikkert, S.E, Zhmak, M.N, Bertrand, D, Tsetlin, V, Sixma, T.K, Smit, A.B. | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Nicotinic Acetylcholine Receptor Homolog Achbp in Complex with an Alpha- Conotoxin Pnia Variant
Nat.Struct.Mol.Biol., 12, 2005
|
|
2BR8
 
 | | Crystal Structure of Acetylcholine-binding Protein (AChBP) from Aplysia californica in complex with an alpha-conotoxin PnIA variant | | Descriptor: | ALPHA-CONOTOXIN PNIA, SOLUBLE ACETYLCHOLINE RECEPTOR, SULFATE ION | | Authors: | Celie, P.H.N, Kasheverov, I.E, Mordvintsev, D.Y, Hogg, R.C, van Nierop, P, van Elk, R, van Rossum-Fikkert, S.E, Zhmak, M.N, Bertrand, D, Tsetlin, V, Sixma, T.K, Smit, A.B. | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Nicotinic Acetylcholine Receptor Homolog Achbp in Complex with an Alpha-Conotoxin Pnia Variant
Nat.Struct.Mol.Biol., 12, 2005
|
|
7RMI
 
 | | SP6-11 biased agonist bound to active human neurokinin 1 receptor in complex with miniGs/q70 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Harris, J.A, Faust, B, Gondin, A.B, Daemgen, M.A, Suomivuori, C.M, Veldhuis, N.A, Cheng, Y, Dror, R.O, Thal, D, Manglik, A. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-03 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Selective G protein signaling driven by substance P-neurokinin receptor dynamics.
Nat.Chem.Biol., 18, 2022
|
|
7RMG
 
 | | Substance P bound to active human neurokinin 1 receptor in complex with miniGs/q70 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Harris, J.A, Faust, B, Gondin, A.B, Daemgen, M.A, Suomivuori, C.M, Veldhuis, N.A, Cheng, Y, Dror, R.O, Thal, D, Manglik, A. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-03 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Selective G protein signaling driven by substance P-neurokinin receptor dynamics.
Nat.Chem.Biol., 18, 2022
|
|
7RMH
 
 | | Substance P bound to active human neurokinin 1 receptor in complex with miniGs399 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Harris, J.A, Faust, B, Gondin, A.B, Daemgen, M.A, Suomivuori, C.M, Veldhuis, N.A, Cheng, Y, Dror, R.O, Thal, D, Manglik, A. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-03 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Selective G protein signaling driven by substance P-neurokinin receptor dynamics.
Nat.Chem.Biol., 18, 2022
|
|
2BJ0
 
 | | Crystal Structure of AChBP from Bulinus truncatus revals the conserved structural scaffold and sites of variation in nicotinic acetylcholine receptors | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ACETYLCHOLINE-BINDING PROTEIN | | Authors: | Celie, P.H.N, Klaassen, R.V, Van Rossum-Fikkert, S.E, Van Elk, R, Van Nierop, P, Smit, A.B, Sixma, T.K. | | Deposit date: | 2005-01-27 | | Release date: | 2005-05-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Acetylcholine-Binding Protein from Bulinus Truncatus Reveals the Conserved Structural Scaffold and Sites of Variation in Nicotinic Acetylcholine Receptors.
J.Biol.Chem., 280, 2005
|
|
2AUX
 
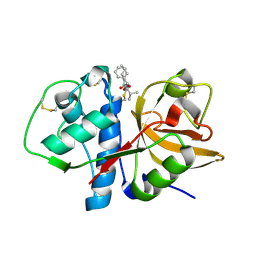 | | Cathepsin K complexed with a semicarbazone inhibitor | | Descriptor: | (1R)-2-METHYL-1-(PHENYLMETHYL)PROPYL[(1S)-1-FORMYLPENTYL]CARBAMATE, Cathepsin K | | Authors: | Adkison, K.K, Barrett, D.G, Deaton, D.N, Gampe, R.T, Hassell, A.M, Long, S.T, McFadyen, R.B, Miller, A.B, Miller, L.R, Shewchuk, L.M. | | Deposit date: | 2005-08-29 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Semicarbazone-based inhibitors of cathepsin K, are they prodrugs for aldehyde inhibitors?
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2C3G
 
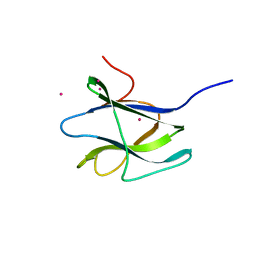 | | Structure of CBM26 from Bacillus halodurans amylase | | Descriptor: | ALPHA-AMYLASE G-6, CADMIUM ION | | Authors: | Boraston, A.B, Healey, M, Klassen, J, Ficko-Blean, E, Lammerts Van Bueren, A, Law, V. | | Deposit date: | 2005-10-07 | | Release date: | 2005-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural and Functional Analysis of Alpha-Glucan Recognition by Family 25 and 26 Carbohydrate-Binding Modules Reveals a Conserved Mode of Starch Recognition
J.Biol.Chem., 281, 2006
|
|
2CNJ
 
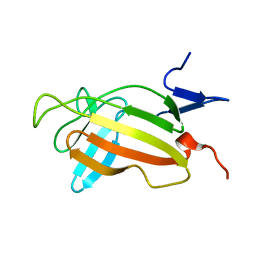 | | NMR studies on the interaction of Insulin-Growth Factor II (IGF-II) with IGF2R domain 11 | | Descriptor: | CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | Authors: | Williams, C, Prince, S, Zaccheo, O, Hassan, A.B, Crosby, J, Crump, M. | | Deposit date: | 2006-05-22 | | Release date: | 2007-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Insights Into the Interaction of Insulin-Like Growth Factor 2 with Igf2R Domain 11.
Structure, 15, 2007
|
|
2C9T
 
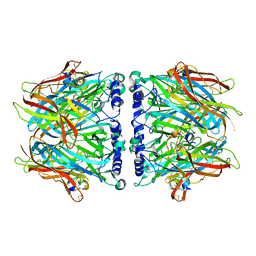 | | Crystal Structure Of Acetylcholine Binding Protein (AChBP) From Aplysia Californica In Complex With alpha-Conotoxin ImI | | Descriptor: | ALPHA-CONOTOXIN IMI, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Hogg, R.C, Celie, P.H, Bertrand, D, Tsetlin, V, Smit, A.B, Sixma, T.K. | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Determinants of Selective {Alpha}-Conotoxin Binding to a Nicotinic Acetylcholine Receptor Homolog Achbp.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2CDO
 
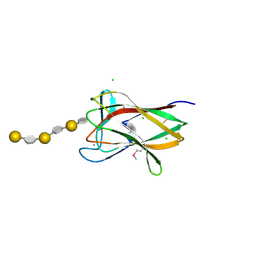 | | structure of agarase carbohydrate binding module in complex with neoagarohexaose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, BETA-AGARASE 1, ... | | Authors: | Henshaw, J, Horne, A, Van Bueren, A.L, Money, V.A, Bolam, D.N, Czjzek, M, Weiner, R.M, Hutcheson, S.W, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2006-01-25 | | Release date: | 2006-02-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Family 6 Carbohydrate Binding Modules in Beta-Agarases Display Exquisite Selectivity for the Non- Reducing Termini of Agarose Chains.
J.Biol.Chem., 281, 2006
|
|
2AUZ
 
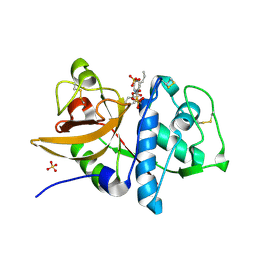 | | Cathepsin K complexed with a semicarbazone inhibitor | | Descriptor: | 1-(PHENYLMETHYL)CYCLOPENTYL[(1S)-1-FORMYLPENTYL]CARBAMATE, Cathepsin K, SULFATE ION | | Authors: | Adkison, K.K, Barrett, D.G, Deaton, D.N, Gampe, R.T, Hassell, A.M, Long, S.T, McFadyen, R.B, Miller, A.B, Miller, L.R, Shewchuk, L.M. | | Deposit date: | 2005-08-29 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Semicarbazone-based inhibitors of cathepsin K, are they prodrugs for aldehyde inhibitors?
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2CDP
 
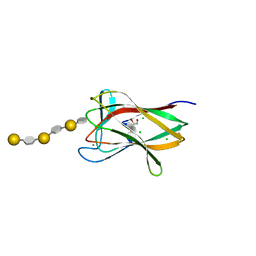 | | Structure of a CBM6 in complex with neoagarohexaose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, BETA-AGARASE 1, ... | | Authors: | Henshaw, J, Horne, A, Van Bueren, A.L, Money, V.A, Bolam, D.N, Czjzek, M, Weiner, R.M, Hutcheson, S.W, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2006-01-26 | | Release date: | 2006-02-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Family 6 Carbohydrate Binding Modules in Beta-Agarases Display Exquisite Selectivity for the Non- Reducing Termini of Agarose Chains.
J.Biol.Chem., 281, 2006
|
|
2BB9
 
 | | Structure of HIV1 protease and AKC4p_133a complex. | | Descriptor: | 2-ETHOXYETHYL (1S,2S)-3-{(2S)-4-[(3AS,8S,8AR)-2-OXO-3,3A,8,8A-TETRAHYDRO-2H-INDENO[1,2-D][1,3]OXAZOL-8-YL]-2-BENZYL-3-OXO-2,3-DIHYDRO-1H-PYRROL-2-YL}-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Smith III, A.B, Charnley, A.K, Kuo, L.C, Munshi, S. | | Deposit date: | 2005-10-17 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design, synthesis, and biological evaluation of monopyrrolinone-based HIV-1 protease inhibitors possessing augmented P2' side chains
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2CGR
 
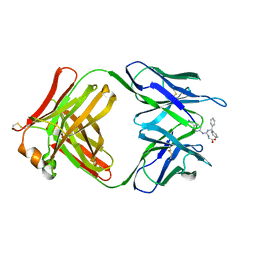 | |
2BBB
 
 | | Structure of HIV1 protease and hh1_173_3a complex. | | Descriptor: | (3S)-TETRAHYDROFURAN-3-YL (1R)-3-{(2R)-4-[(1S,3S)-3-(2-AMINO-2-OXOETHYL)-2,3-DIHYDRO-1H-INDEN-1-YL]-2-BENZYL-3-OXO-2,3-DIHYDRO-1H-PYRROL-2-YL}-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Smith III, A.B, Charnley, A.K, Kuo, L.C, Munshi, S. | | Deposit date: | 2005-10-17 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, synthesis, and biological evaluation of monopyrrolinone-based HIV-1 protease inhibitors possessing augmented P2' side chains
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2CG4
 
 | | Structure of E.coli AsnC | | Descriptor: | ASPARAGINE, MAGNESIUM ION, REGULATORY PROTEIN ASNC | | Authors: | Thaw, P, Sedelnikova, S.E, Muranova, T, Wiese, S, Ayora, S, Alonso, J.C, Brinkman, A.B, Akerboom, J, van der Oost, J, Rafferty, J.B. | | Deposit date: | 2006-02-27 | | Release date: | 2006-04-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight Into Gene Transcriptional Regulation and Effector Binding by the Lrp/Asnc Family.
Nucleic Acids Res., 34, 2006
|
|
1UUT
 
 | | The Nuclease Domain of Adeno-Associated Virus Rep Complexed with the RBE' Stemloop of the Viral Inverted Terminal Repeat | | Descriptor: | 5'-D(*CP*AP*GP*CP*TP*CP*TP*TP*TP*GP *AP*GP*CP*TP*G)-3', CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Dyda, F, Hickman, A.B, Ronning, D.R, Perez, Z.N, Kotin, R.M. | | Deposit date: | 2004-01-10 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Nuclease Domain of Adeno-Associated Virus Rep Coordinates Replication Initiation Using Two Distinct DNA Recognition Interfaces
Mol.Cell, 13, 2004
|
|
1UY4
 
 | |
