1CGS
 
 | |
4EGO
 
 | | The X-ray crystal structure of CYP199A4 in complex with indole-6-carboxylic acid | | Descriptor: | 1H-indole-6-carboxylic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Zhou, R.M, Tan, A.B.H, Wong, L.-L. | | Deposit date: | 2012-03-31 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Investigation of the substrate range of CYP199A4: modification of the partition between hydroxylation and desaturation activities by substrate and protein engineering
Chemistry, 18, 2012
|
|
4EGM
 
 | | The X-ray crystal structure of CYP199A4 in complex with 4-ethylbenzoic acid | | Descriptor: | 4-ethylbenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Zhou, R.M, Tan, A.B.H, Wong, L.-L. | | Deposit date: | 2012-03-31 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Investigation of the substrate range of CYP199A4: modification of the partition between hydroxylation and desaturation activities by substrate and protein engineering
Chemistry, 18, 2012
|
|
3MHH
 
 | | Structure of the SAGA Ubp8/Sgf11/Sus1/Sgf73 DUB module | | Descriptor: | Protein SUS1, SAGA-associated factor 11, SAGA-associated factor 73, ... | | Authors: | Samara, N.L, Datta, A.B, Berndsen, C.E, Zhang, X, Yao, T, Cohen, R.E, Wolberger, C. | | Deposit date: | 2010-04-08 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into the assembly and function of the SAGA deubiquitinating module.
Science, 328, 2010
|
|
3LZN
 
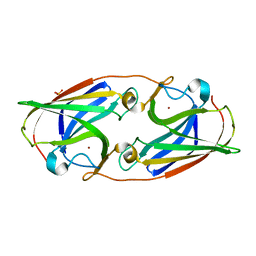 | | Crystal Structure Analysis of the apo P19 protein from Campylobacter jejuni at 1.59 A at pH 9 | | Descriptor: | P19 protein, SULFATE ION, ZINC ION | | Authors: | Doukov, T.I, Chan, A.C.K, Scofield, M, Ramin, A.B, Tom-Yew, S.A.L, Murphy, M.E.P. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure and Function of P19, a High-Affinity Iron Transporter of the Human Pathogen Campylobacter jejuni.
J.Mol.Biol., 401, 2010
|
|
2KTU
 
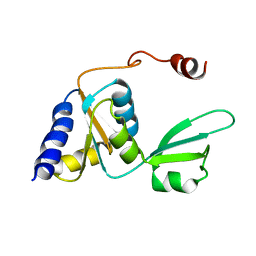 | |
3LZP
 
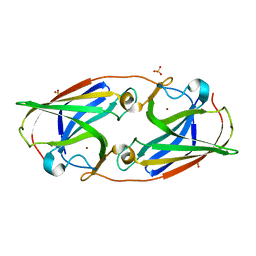 | | Crystal Structure Analysis of the 'as-isolated' P19 protein from Campylobacter jejuni at 1.65 A at pH 9.0 | | Descriptor: | COPPER (II) ION, P19 protein, SULFATE ION | | Authors: | Doukov, T.I, Chan, A.C.K, Scofield, M, Ramin, A.B, Tom-Yew, S.A.L, Murphy, M.E.P. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Function of P19, a High-Affinity Iron Transporter of the Human Pathogen Campylobacter jejuni.
J.Mol.Biol., 401, 2010
|
|
4EGN
 
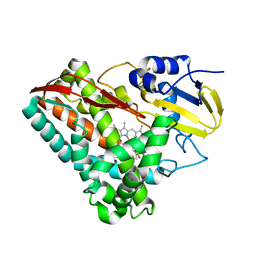 | | The X-ray crystal structure of CYP199A4 in complex with veratric acid | | Descriptor: | 3,4-dimethoxybenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Zhou, R.M, Tan, A.B.H, Wong, L.-L. | | Deposit date: | 2012-03-31 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigation of the substrate range of CYP199A4: modification of the partition between hydroxylation and desaturation activities by substrate and protein engineering
Chemistry, 18, 2012
|
|
2MUN
 
 | |
1A8J
 
 | |
3UAQ
 
 | | Crystal Structure of the N-lobe Domain of Lactoferrin Binding Protein B (LbpB) of Moraxella bovis | | Descriptor: | LbpB B-lobe | | Authors: | Arutyunova, E, Brooks, C.L, Beddeck, A, Mak, M.W, Schryvers, A.B, Lemieux, M.J. | | Deposit date: | 2011-10-21 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9318 Å) | | Cite: | Crystal structure of the N-lobe of lactoferrin binding protein B from Moraxella bovis(1).
Biochem.Cell Biol., 90, 2012
|
|
4HPM
 
 | | PCGF1 Ub fold (RAWUL)/BCORL1 PUFD Complex | | Descriptor: | BCL-6 corepressor-like protein 1, PHOSPHATE ION, Polycomb group RING finger protein 1 | | Authors: | Junco, S.E, Wang, R, Gaipa, J, Taylor, A.B, Gearhart, M.D, Bardwell, V.J, Hart, P.J, Kim, C.A. | | Deposit date: | 2012-10-24 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the Polycomb Group Protein PCGF1 in Complex with BCOR Reveals Basis for Binding Selectivity of PCGF Homologs.
Structure, 21, 2013
|
|
3KSQ
 
 | | Discovery of C-Imidazole Azaheptapyridine FPT Inhibitors | | Descriptor: | FARNESYL DIPHOSPHATE, Farnesyltransferase, CAAX box, ... | | Authors: | Zhu, H.Y, Cooper, A.B, Njoroge, G, Kirschmeier, P, Strickland, C, Hruza, A, Girijavallabhan, V.M. | | Deposit date: | 2009-11-23 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of C-imidazole azaheptapyridine FPT inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4FUP
 
 | |
3L4K
 
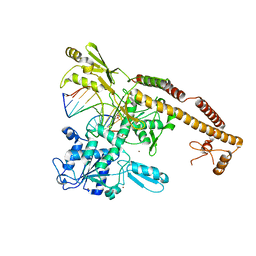 | | Topoisomerase II-DNA cleavage complex, metal-bound | | Descriptor: | 3'-THIO-THYMIDINE-5'-PHOSPHATE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*CP*GP*TP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*CP*GP*CP*GP*GP*TP*AP*GP*CP*AP*GP*TP*AP*GP*G)-3'), ... | | Authors: | Schmidt, B.H, Burgin, A.B, Deweese, J.E, Osheroff, N, Berger, J.M. | | Deposit date: | 2009-12-20 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases.
Nature, 465, 2010
|
|
1BIU
 
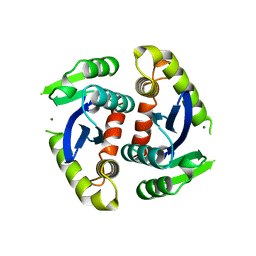 | | HIV-1 INTEGRASE CORE DOMAIN COMPLEXED WITH MG++ | | Descriptor: | HIV-1 INTEGRASE, MAGNESIUM ION | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-19 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
2MCG
 
 | |
1J83
 
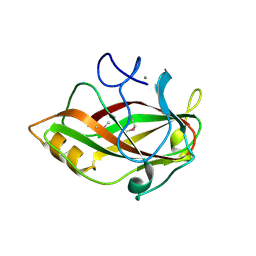 | | STRUCTURE OF FAM17 CARBOHYDRATE BINDING MODULE FROM CLOSTRIDIUM CELLULOVORANS | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA GLUCANASE ENGF | | Authors: | Notenboom, V, Boraston, A.B, Chiu, P, Freelove, A.C.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-05-20 | | Release date: | 2001-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition of cello-oligosaccharides by a family 17 carbohydrate-binding module: an X-ray crystallographic, thermodynamic and mutagenic study.
J.Mol.Biol., 314, 2001
|
|
4FUN
 
 | |
1J84
 
 | | STRUCTURE OF FAM17 CARBOHYDRATE BINDING MODULE FROM CLOSTRIDIUM CELLULOVORANS WITH BOUND CELLOTETRAOSE | | Descriptor: | CALCIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, endo-1,4-beta glucanase EngF | | Authors: | Notenboom, V, Boraston, A.B, Chiu, P, Freelove, A.C.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-05-20 | | Release date: | 2001-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Recognition of cello-oligosaccharides by a family 17 carbohydrate-binding module: an X-ray crystallographic, thermodynamic and mutagenic study.
J.Mol.Biol., 314, 2001
|
|
1BJP
 
 | | CRYSTAL STRUCTURE OF 4-OXALOCROTONATE TAUTOMERASE INACTIVATED BY 2-OXO-3-PENTYNOATE AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | 2-OXO-3-PENTENOIC ACID, 4-OXALOCROTONATE TAUTOMERASE | | Authors: | Taylor, A.B, Czerwinski, R.M, Johnson Junior, W.H, Whitman, C.P, Hackert, M.L. | | Deposit date: | 1998-06-26 | | Release date: | 1998-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of 4-oxalocrotonate tautomerase inactivated by 2-oxo-3-pentynoate at 2.4 A resolution: analysis and implications for the mechanism of inactivation and catalysis.
Biochemistry, 37, 1998
|
|
1G6O
 
 | | CRYSTAL STRUCTURE OF THE HELICOBACTER PYLORI ATPASE, HP0525, IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CAG-ALPHA, DI(HYDROXYETHYL)ETHER | | Authors: | Yeo, H.J, Savvides, S.N, Herr, A.B, Lanka, E, Waksman, G, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-11-07 | | Release date: | 2001-01-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system.
Mol.Cell, 6, 2000
|
|
4FUM
 
 | |
1FXZ
 
 | | CRYSTAL STRUCTURE OF SOYBEAN PROGLYCININ A1AB1B HOMOTRIMER | | Descriptor: | GLYCININ G1 | | Authors: | Adachi, M, Takenaka, Y, Gidamis, A.B, Mikami, B, Utsumi, S. | | Deposit date: | 2000-09-28 | | Release date: | 2001-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of soybean proglycinin A1aB1b homotrimer.
J.Mol.Biol., 305, 2001
|
|
2KTV
 
 | |
