5FSP
 
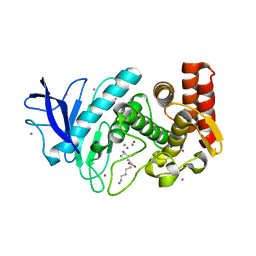 | | Structure of thermolysin prepared by the 'soak-and-freeze' method under 100 bar of krypton pressure | | Descriptor: | CALCIUM ION, KRYPTON, LYSINE, ... | | Authors: | Lafumat, B, Mueller-Dieckmann, C, Colloc'h, N, Prange, T, Royant, A, van der Linden, p, Carpentier, P. | | Deposit date: | 2016-01-06 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Gas-Sensitive Biological Crystals Processed in Pressurized Oxygen and Krypton Atmospheres: Deciphering Gas Channels in Proteins Using a Novel `Soak-and-Freeze' Methodology.
J.Appl.Crystallogr., 49, 2016
|
|
7KXO
 
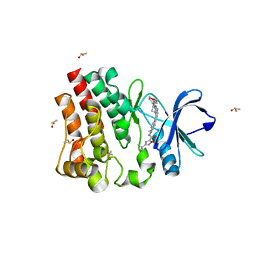 | | BTK1 SOAKED WITH COMPOUND 24 | | Descriptor: | 3-tert-butyl-N-[(1R)-6-{2-[5-methyl-1-(oxan-4-yl)-1H-pyrazol-4-yl]-3H-imidazo[4,5-b]pyridin-7-yl}-1,2,3,4-tetrahydronaphthalen-1-yl]-1,2,4-oxadiazole-5-carboxamide, DIMETHYL SULFOXIDE, Isoform BTK-C of Tyrosine-protein kinase BTK | | Authors: | Viacava Follis, A. | | Deposit date: | 2020-12-04 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of potent and selective reversible Bruton's tyrosine kinase inhibitors.
Bioorg.Med.Chem., 40, 2021
|
|
5AG7
 
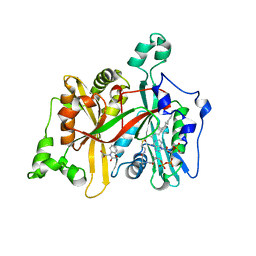 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A BENZOMORPHOLINE LIGAND | | Descriptor: | GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA, ethyl (3-oxo-2,3-dihydro-4H-1,4-benzoxazin-4-yl)acetate | | Authors: | Robinson, D.A, Spinks, D, Smith, V.C, Thompson, S, Smith, A, Torrie, L.S, McElroy, S.P, Brand, S, Brenk, R, Frearson, J.A, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2015-01-29 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development of Small-Molecule Trypanosoma Brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode.
Chemmedchem, 10, 2015
|
|
6C64
 
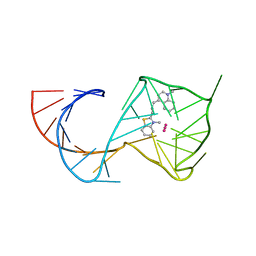 | | Crystal Structure of the Mango-II Fluorescent Aptamer Bound to TO3-Biotin | | Descriptor: | 1-methyl-4-[(1E)-3-(3-methyl-1,3-benzothiazol-3-ium-2-yl)prop-1-en-1-yl]quinolin-1-ium, POTASSIUM ION, RNA (32-MER), ... | | Authors: | Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2018-01-17 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.00014877 Å) | | Cite: | Crystal Structures of the Mango-II RNA Aptamer Reveal Heterogeneous Fluorophore Binding and Guide Engineering of Variants with Improved Selectivity and Brightness.
Biochemistry, 57, 2018
|
|
7KXP
 
 | | BTK1 SOAKED WITH COMPOUND 25 | | Descriptor: | 3-tert-butyl-N-[(1S)-6-{2-[3-methyl-1-(oxan-4-yl)-1H-pyrazol-4-yl]-1H-imidazo[4,5-b]pyridin-7-yl}-1,2,3,4-tetrahydronaphthalen-1-yl]-1,2,4-oxadiazole-5-carboxamide, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Viacava Follis, A. | | Deposit date: | 2020-12-04 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of potent and selective reversible Bruton's tyrosine kinase inhibitors.
Bioorg.Med.Chem., 40, 2021
|
|
5AE5
 
 | | Structures of inactive and activated DntR provide conclusive evidence for the mechanism of action of LysR transcription factors | | Descriptor: | GLYCEROL, LYSR-TYPE REGULATORY PROTEIN | | Authors: | Lerche, M, Dian, C, Round, A, Lonneborg, R, Brzezinski, P, Leonard, G.A. | | Deposit date: | 2015-08-25 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | The Solution Configurations of Inactive and Activated Dntr Have Implications for the Sliding Dimer Mechanism of Lysr Transcription Factors.
Sci.Rep., 6, 2016
|
|
6WVS
 
 | | Hyperstable de novo TIM barrel variant DeNovoTIM15 | | Descriptor: | DeNovoTIM15 hyperstable de novo TIM barrel | | Authors: | Bick, M.J, Haydon, I.C, Caldwell, S.J, Zeymer, C, Huang, P, Fernandez-Velasco, D.A, Baker, D. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Tight and specific lanthanide binding in a de novo TIM barrel with a large internal cavity designed by symmetric domain fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8OT2
 
 | | Structural and functional studies of geldanamycin amide synthase ShGdmF | | Descriptor: | 1,2-ETHANEDIOL, 3-azanyl-5-methyl-phenol, ACETATE ION, ... | | Authors: | Ewert, W, Zeilinger, C, Kirschning, A, Preller, M. | | Deposit date: | 2023-04-20 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and functional studies of geldanamycin amide synthase ShGdmF
To Be Published
|
|
6WW5
 
 | | Structure of VcINDY-Na-Fab84 in nanodisc | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, DASS family sodium-coupled anion symporter, Fab84 Heavy Chain, ... | | Authors: | Sauer, D.B, Marden, J, Song, J.M, Koide, A, Koide, S, Wang, D.N. | | Deposit date: | 2020-05-07 | | Release date: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
4UEU
 
 | | Tyrosine kinase AS - a common ancestor of Src and Abl | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, TYROSINE KINASE AS - A COMMON ANCESTOR OF SRC AND ABL | | Authors: | Kutter, S, Wilson, C, Agafonov, R.V, Hoemberger, M.S, Zorba, A, Halpin, J.C, Theobald, D.L, Kern, D. | | Deposit date: | 2014-12-18 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Kinase Dynamics. Using Ancient Protein Kinases to Unravel a Modern Cancer Drug'S Mechanism.
Science, 347, 2015
|
|
5FLW
 
 | | Crystal structure of putative exo-beta-1,3-galactanase from Bifidobacterium bifidum s17 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, EXO-BETA-1,3-GALACTANASE | | Authors: | Godoy, A.S, de Lima, M.Z.T, Ramia, M.P, Camilo, C.M, Muniz, J.R.C, Polikarpov, I. | | Deposit date: | 2015-10-28 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Crystal structure of a putative exo-beta-1,3-galactanase from Bifidobacterium bifidum S17.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5JI3
 
 | | HslUV complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV | | Authors: | Grant, R.A, Sauer, R.T, Schmitz, K.R, Baytshtok, V. | | Deposit date: | 2016-04-21 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Structurally Dynamic Region of the HslU Intermediate Domain Controls Protein Degradation and ATP Hydrolysis.
Structure, 24, 2016
|
|
5JJJ
 
 | | Structure of the SRII/HtrII Complex in P64 space group ("U" shape) | | Descriptor: | EICOSANE, RETINAL, Sensory rhodopsin II transducer, ... | | Authors: | Ishchenko, A, Round, E, Borshchevskiy, V, Grudinin, S, Gushchin, I, Klare, J, Remeeva, A, Polovinkin, V, Utrobin, P, Balandin, T, Engelhard, M, Bueldt, G, Gordeliy, V. | | Deposit date: | 2016-04-24 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New Insights on Signal Propagation by Sensory Rhodopsin II/Transducer Complex.
Sci Rep, 7, 2017
|
|
6G1H
 
 | | Amine Dehydrogenase from Petrotoga mobilis; open form | | Descriptor: | 1,2-ETHANEDIOL, Dihydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Beloti, L, Frese, A, Mayol, O, Vergne-Vaxelaire, C, Grogan, G. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nat Catal, 2019
|
|
5FTZ
 
 | | AA10 lytic polysaccharide monooxygenase (LPMO) from Streptomyces lividans | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION | | Authors: | Chaplin, A.K.C, Wilson, M.T, Hough, M.A, Svistunenko, D.A, Hemsworth, G.R, Walton, P.H, Vijgenboom, E, Worrall, J.A.R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Heterogeneity in the Histidine-Brace Copper Coordination Sphere in Aa10 Lytic Polysaccharide Monooxygenases.
J.Biol.Chem., 291, 2016
|
|
5FYM
 
 | | Crystal structure of JmjC domain of human histone demethylase UTY in complex with D-2-hydroxyglutarate | | Descriptor: | (2R)-2-hydroxypentanedioic acid, 1,2-ETHANEDIOL, HISTONE DEMETHYLASE UTY, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Jmjc Domain of Human Histone Demethylase Uty in Complex with D-2- Hydroxyglutarate
To be Published
|
|
1A2C
 
 | | Structure of thrombin inhibited by AERUGINOSIN298-A from a BLUE-GREEN ALGA | | Descriptor: | Aeruginosin 298-A, Hirudin variant-2, SODIUM ION, ... | | Authors: | Rios-Steiner, J.L, Murakami, M, Tulinsky, A. | | Deposit date: | 1997-12-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Thrombin Inhibited by Aeruginosin 298-A from a Blue-Green Alga
J.Am.Chem.Soc., 120, 1998
|
|
7KZ6
 
 | | Crystal structure of KabA from Bacillus cereus UW85 with bound cofactor PMP | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme | | Authors: | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
1A4M
 
 | | ADA STRUCTURE COMPLEXED WITH PURINE RIBOSIDE AT PH 7.0 | | Descriptor: | 6-HYDROXY-1,6-DIHYDRO PURINE NUCLEOSIDE, ADENOSINE DEAMINASE, ZINC ION | | Authors: | Wang, Z, Quiocho, F.A. | | Deposit date: | 1998-01-31 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Complexes of adenosine deaminase with two potent inhibitors: X-ray structures in four independent molecules at pH of maximum activity.
Biochemistry, 37, 1998
|
|
8P1S
 
 | | Bifidobacterium asteroides alpha-L-fucosidase (TT1819) in complex with fucose. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifidobacterium asteroides alpha-L-fucosidase (TT1819), ... | | Authors: | Owen, C.D, Penner, M, Gascuena, A.N, Wu, H, Hernando, P.J, Monaco, S, Le Gall, G, Gardner, R, Ndeh, D, Urbanowicz, P.A, Spencer, D.I.R, Walsh, M.A, Angulo, J, Juge, N. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring sequence, structure and function of microbial fucosidases from glycoside hydrolase GH29 family
To Be Published
|
|
4UNQ
 
 | | Mtb TMK in complex with compound 36 | | Descriptor: | 4-[(R)-methylsulfinyl]-2-oxo-6-[3-(trifluoromethoxy)phenyl]-1,2-dihydropyridine-3-carbonitrile, SODIUM ION, THYMIDYLATE KINASE | | Authors: | Read, J.A, Hussein, S, Gingell, H, Tucker, J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure Guided Lead Generation for M. Tuberculosis Thymidylate Kinase (Mtb Tmk): Discovery of 3-Cyanopyridone and 1,6-Naphthyridin-2-One as Potent Inhibitors.
J.Med.Chem., 58, 2015
|
|
4UMI
 
 | |
6G3P
 
 | |
6G47
 
 | | Crystal Structure of Human Adenovirus 52 Short Fiber Knob in Complex with alpha-(2,8)-Trisialic Acid (DP3) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Liaci, A.M, Stehle, T. | | Deposit date: | 2018-03-26 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Polysialic acid is a cellular receptor for human adenovirus 52.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C6T
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound with RfaH | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Artsimovitch, I, Landick, R, Darst, S.A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators.
Cell, 173, 2018
|
|
