2IES
 
 | |
5CTV
 
 | | Catalytic domain of LytA, the major autolysin of Streptococcus pneumoniae, (C60A, H133A, C136A mutant) complexed with peptidoglycan fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, Autolysin, fragment of peptidoglycan | | Authors: | Achour, A, Sandalova, T, Mellroth, P. | | Deposit date: | 2015-07-24 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The crystal structure of the major pneumococcal autolysin LytA in complex with a large peptidoglycan fragment reveals the pivotal role of glycans for lytic activity.
Mol.Microbiol., 101, 2016
|
|
2J4Z
 
 | | Structure of Aurora-2 in complex with PHA-680626 | | Descriptor: | 4-(4-METHYLPIPERAZIN-1-YL)-N-[5-(2-THIENYLACETYL)-1,5-DIHYDROPYRROLO[3,4-C]PYRAZOL-3-YL]BENZAMIDE, ARSENIC, SERINE THREONINE-PROTEIN KINASE 6 | | Authors: | Cameron, A.D, Izzo, G, Storici, P, Rusconi, L, Fancelli, D, Varasi, M, Berta, D, Bindi, S, Forte, B, Severino, D, Tonani, R, Vianello, P. | | Deposit date: | 2006-09-08 | | Release date: | 2006-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1,4,5,6-tetrahydropyrrolo[3,4-c]pyrazoles: identification of a potent Aurora kinase inhibitor with a favorable antitumor kinase inhibition profile.
J. Med. Chem., 49, 2006
|
|
1JIN
 
 | | P450eryF/ketoconazole | | Descriptor: | CIS-1-ACETYL-4-(4-((2-(2,4-DICHLOROPHENYL)-2-(1H-IMIDAZOL-1-YLMETHYL)-1,3-DIOXOLAN-4-YL)METHOXY)PHENYL)PIPERAZINE, CYTOCHROME P450 107A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cupp-Vickery, J.R, Garcia, C, Hofacre, A, McGee-Estrada, K. | | Deposit date: | 2001-07-02 | | Release date: | 2001-10-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ketoconazole-induced conformational changes in the active site of cytochrome P450eryF.
J.Mol.Biol., 311, 2001
|
|
5V26
 
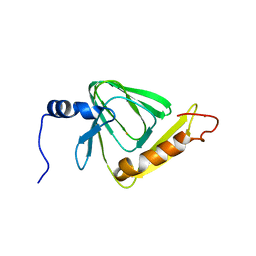 | |
7ZT6
 
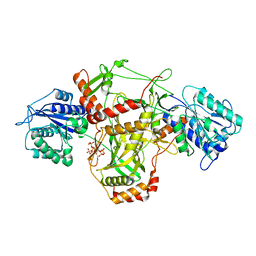 | |
5VAH
 
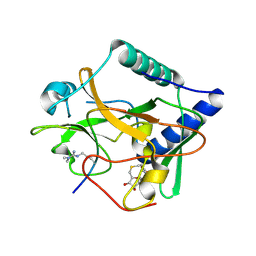 | | Crystal structure of ATXR5 SET domain in complex with histone H3 di-methylated on R26 | | Descriptor: | Histone H3.2, Probable Histone-lysine N-methyltransferase ATXR5, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.-F. | | Deposit date: | 2017-03-26 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
2INS
 
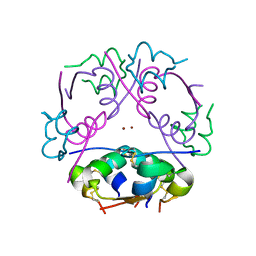 | | THE STRUCTURE OF DES-PHE B1 BOVINE INSULIN | | Descriptor: | DES-PHE B1 INSULIN (CHAIN A), DES-PHE B1 INSULIN (CHAIN B), ZINC ION | | Authors: | Smith, G.D, Duax, W.L, Dodson, E.J, Dodson, G.G, Degraaf, R.A.G, Reynolds, C.D. | | Deposit date: | 1982-05-10 | | Release date: | 1982-08-05 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of Des-Phe B1 Bovine Insulin
Acta Crystallogr.,Sect.B, 38, 1982
|
|
3ZDX
 
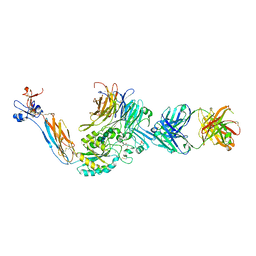 | | Integrin alphaIIB beta3 headpiece and RGD peptide complex | | Descriptor: | 10E5 FAB, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Zhu, J.H, Zhu, J.Q, Springer, T.A. | | Deposit date: | 2012-12-03 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Complete Integrin Headpiece Opening in Eight Steps.
J.Cell Biol., 201, 2013
|
|
7AWV
 
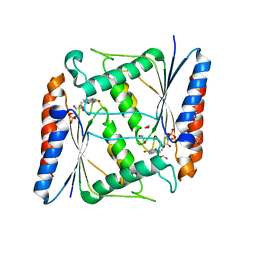 | | Azoreductase (AzoRo) from Rhodococcus opacus 1CP | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase, ... | | Authors: | Bento, I, Ngo, A, Qi, J, Juric, C, Tischler, D. | | Deposit date: | 2020-11-09 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of molecular basis that underlie enzymatic specificity of AzoRo from Rhodococcus opacus 1CP: A potential NADH:quinone oxidoreductase.
Arch.Biochem.Biophys., 717, 2022
|
|
2W5R
 
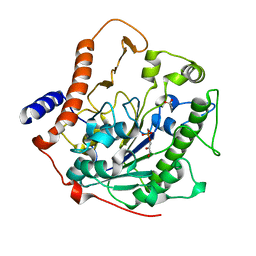 | | Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS. | | Descriptor: | (2R)-2,3-dihydroxypropyl phosphate, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Lu, D, Wormann, M.E, Zhang, X, Schneewind, O, Grundling, A, Freemont, P.S. | | Deposit date: | 2008-12-11 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Mechanism of Lipoteichoic Acid Synthesis by Staphylococcus Aureus Ltas.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ILZ
 
 | |
1TGN
 
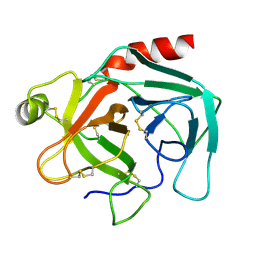 | |
4RCF
 
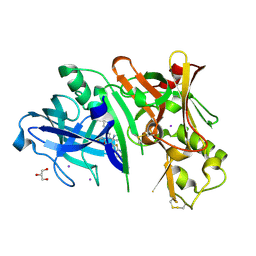 | | Crystal structure of BACE1 in complex with 2-aminooxazoline 4-fluoroxanthene inhibitor 49 | | Descriptor: | (4S)-2'-(3,6-dihydro-2H-pyran-4-yl)-4'-fluoro-7'-(2-fluoropyridin-3-yl)spiro[1,3-oxazole-4,9'-xanthen]-2-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-24 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Lead Optimization and Modulation of hERG Activity in a Series of Aminooxazoline Xanthene beta-Site Amyloid Precursor Protein Cleaving Enzyme (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
4QXW
 
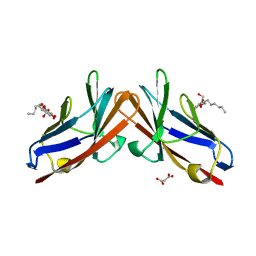 | | Crystal structure of the human CEACAM1 membrane distal amino terminal (N)-domain | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, MALONIC ACID, octyl beta-D-glucopyranoside | | Authors: | Huang, Y.H, Gandhi, A.K, Russell, A, Kondo, Y, Chen, Q, Petsko, G.A, Blumberg, R.S. | | Deposit date: | 2014-07-22 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | CEACAM1 regulates TIM-3-mediated tolerance and exhaustion.
Nature, 517, 2015
|
|
5KI5
 
 | | Structural impact of single ribonucleotides in DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*CP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*CP*CP*TP*G)-3') | | Authors: | Evich, M, Spring-Connell, A.M, Storici, F, Germann, M.W. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Impact of Single Ribonucleotide Residues in DNA.
Chembiochem, 17, 2016
|
|
5KIE
 
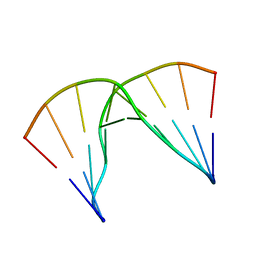 | | Structural impact of single ribonucleotides in DNA | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*TP*CP*CP*AP*T)-3'), DNA/RNA (5'-D(*AP*TP*GP*GP*A)-R(P*G)-D(P*CP*TP*C)-3') | | Authors: | Evich, M, Spring-Connell, A.M, Storici, F, Germann, M.W. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Impact of Single Ribonucleotide Residues in DNA.
Chembiochem, 17, 2016
|
|
5KDP
 
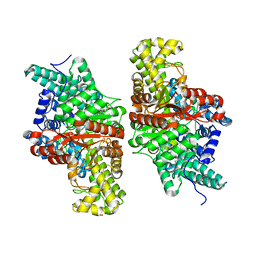 | | E491A mutant of choline TMA-lyase | | Descriptor: | Choline trimethylamine-lyase, MALONATE ION, SODIUM ION | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2016-06-08 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis of C-N Bond Cleavage by the Glycyl Radical Enzyme Choline Trimethylamine-Lyase.
Cell Chem Biol, 23, 2016
|
|
7ZMY
 
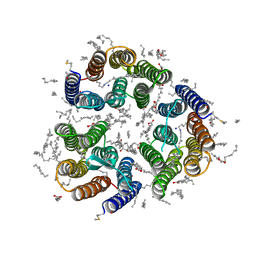 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the ground state at pH 8.2 in the presence of sodium at 100K | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6VQY
 
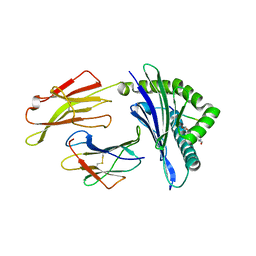 | | HLA-B*27:05 presenting an HIV-1 7mer peptide | | Descriptor: | 7-mer peptide, ARGININE, Beta-2-microglobulin, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
7ZN0
 
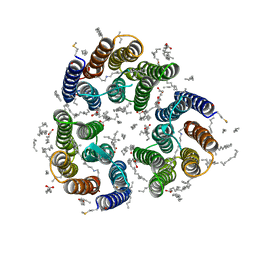 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the M state at pH 8.2 in the presence of sodium at 100K | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3ZGY
 
 | |
7ZN3
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the L state at pH 8.2 in the presence of sodium at 100K | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZN9
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the M state at pH 7.0 in the presence of sodium at 100K | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZNB
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the M state at pH 5.2 in the presence of sodium at 100K | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
