8GDT
 
 | |
8GV1
 
 | | Crystal structure of anti-FX IgG fab with FAST-Ig mutations | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Anti-factor X IgG fab heavy chain, ... | | Authors: | Koga, H, Yamano, T, Fukami, T.A, Sampei, Z, Shiraiwa, H, Torizawa, T. | | Deposit date: | 2022-09-14 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.186 Å) | | Cite: | Efficient production of bispecific antibody by FAST-Ig TM and its application to NXT007 for the treatment of hemophilia A.
Mabs, 15, 2023
|
|
8GV2
 
 | | Crystal structure of anti-FX IgG fab without FAST-Ig mutations | | Descriptor: | 1,2-ETHANEDIOL, Anti-factor X IgG fab heavy chain, Anti-factor X IgG fab light chain | | Authors: | Koga, H, Yamano, T, Fukami, T.A, Sampei, Z, Shiraiwa, H, Torizawa, T. | | Deposit date: | 2022-09-14 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.274 Å) | | Cite: | Efficient production of bispecific antibody by FAST-Ig TM and its application to NXT007 for the treatment of hemophilia A.
Mabs, 15, 2023
|
|
8BCI
 
 | |
8BEG
 
 | | Structure of Ig-like domains from PrgB | | Descriptor: | MAGNESIUM ION, PrgB | | Authors: | Jarva, M, Schmitt, A, Berntsson, R.P.-A. | | Deposit date: | 2022-10-21 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural foundation for the role of enterococcal PrgB in conjugation, biofilm formation, and virulence.
Elife, 12, 2023
|
|
8GUZ
 
 | | Crystal structure of anti-FIXa IgG fab with FAST-Ig mutations | | Descriptor: | 1,2-ETHANEDIOL, Anti-factor IXa IgG fab heavy chain, Anti-factor IXa IgG fab light chain | | Authors: | Koga, H, Yamano, T, Fukami, T.A, Sampei, Z, Shiraiwa, H, Torizawa, T. | | Deposit date: | 2022-09-14 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Efficient production of bispecific antibody by FAST-Ig TM and its application to NXT007 for the treatment of hemophilia A.
Mabs, 15, 2023
|
|
5I06
 
 | | tRNA-guanine Transglycosylase (TGT) in co-crystallized complex with 6-Amino-4-[2-(4-methoxyphenyl)ethyl]-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 1,2-ETHANEDIOL, 6-AMINO-4-[2-(4-METHOXYPHENYL)ETHYL]-1,7-DIHYDRO-8H-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, GLYCEROL, ... | | Authors: | Ehrmann, F.R, Heine, A, Klebe, G. | | Deposit date: | 2016-02-03 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Co-crystallization, Isothermal titration calorimetry and nanoESI-MS reveal dimer disturbing inhibitors
To be Published
|
|
8BAO
 
 | |
6H6S
 
 | | Sad phasing on nickel-substituted human carbonic anhydrase II | | Descriptor: | Carbonic anhydrase 2, NICKEL (II) ION | | Authors: | Calderone, V, Fragai, M, Silva, J.P, Luchinat, C, Ravera, E, Geraldes, C.F.G.C, Macedo, A.L, Cerofolini, L, Giuntini, S. | | Deposit date: | 2018-07-30 | | Release date: | 2019-01-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Non-crystallographic symmetry in proteins: Jahn-Teller-like and Butterfly-like effects?
J. Biol. Inorg. Chem., 24, 2019
|
|
5I1G
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV3-53/IGKV3-11 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, SULFATE ION | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
6SE3
 
 | | Crystal Structure of Ancestral Flavin-containing monooxygenase (FMO) 3-6 | | Descriptor: | Ancestral Flavin-containing monooxygenase (FMO) 3-6, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Nicoll, C, Bailleul, G, Fiorentini, F, Mascotti, M.L, Fraaije, M, Mattevi, A. | | Deposit date: | 2019-07-29 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ancestral-sequence reconstruction unveils the structural basis of function in mammalian FMOs.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8DVM
 
 | | Crystal structure of LRP6 E3E4 in complex with disulfide constrained peptide E3.6 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thakur, A.K, Liau, N.P.D, Sudhamsu, J, Hannoush, R.N. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthetic Multivalent Disulfide-Constrained Peptide Agonists Potentiate Wnt1/ beta-Catenin Signaling via LRP6 Coreceptor Clustering.
Acs Chem.Biol., 18, 2023
|
|
8QQ1
 
 | | SpNOX dehydrogenase domain, mutant F397W in complex with Flavin adenine dinucleotide (FAD) | | Descriptor: | BROMIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Oxidoreductase | | Authors: | Humm, A.S, Dupeux, F, Vermot, A, Petit-Harleim, I, Fieschi, F, Marquez, J.A. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | X-ray structure and enzymatic study of a bacterial NADPH oxidase highlight the activation mechanism of eukaryotic NOX.
Elife, 13, 2024
|
|
6H0X
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand AA040 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | Deposit date: | 2018-07-10 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure of KDM4D with tetrazolylhydrazide ligand AA040
To be published
|
|
7JGN
 
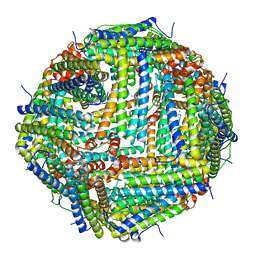 | | Crystal Structure of the Zn-bound Human Heavy-chain variant 122H-delta C-star with meta-benzenedihyrdoxamate collected at 100K | | Descriptor: | Ferritin heavy chain, N~1~,N~3~-dihydroxybenzene-1,3-dicarboxamide, SODIUM ION, ... | | Authors: | Bailey, J.B, Tezcan, F.A. | | Deposit date: | 2020-07-19 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Tunable and Cooperative Thermomechanical Properties of Protein-Metal-Organic Frameworks.
J.Am.Chem.Soc., 142, 2020
|
|
7JFQ
 
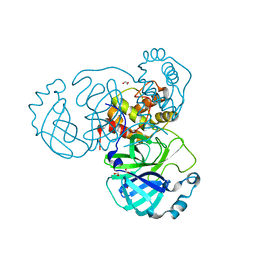 | | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, FORMIC ACID | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-17 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145
To Be Published
|
|
5AFZ
 
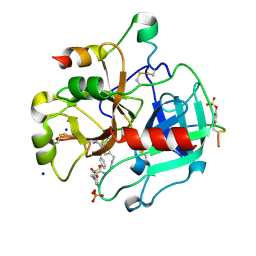 | | Thrombin in complex with (2R)-2-(benzylsulfonylamino)-N-(2-((4- carbamimidoylphenyl)methylamino)-2-oxo-propyl)-3-phenyl-propanamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin-2, N-(BENZYLSULFONYL)-D-PHENYLALANYL-N-(4-CARBAMIMIDOYLBENZYL)GLYCINAMIDE, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2015-01-27 | | Release date: | 2015-08-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Fragments Can Bind Either More Enthalpy or Entropy-Driven: Crystal Structures and Residual Hydration Pattern Suggest Why.
J.Med.Chem., 58, 2015
|
|
7N0J
 
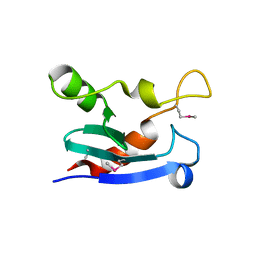 | | Structure of YebY from E. coli K12 | | Descriptor: | YebY | | Authors: | Hadley, R.C, Rosenzweig, A.C. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The copper-linked Escherichia coli AZY operon: Structure, metal binding, and a possible physiological role in copper delivery.
J.Biol.Chem., 298, 2022
|
|
4MQD
 
 | | Crystal structure of ComJ, inhibitor of the DNA degrading activity of NucA, from Bacillus subtilis | | Descriptor: | DNA-entry nuclease inhibitor | | Authors: | Chang, C, Mack, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-16 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of ComJ, inhibitor of the DNA degrading activity of NucA, from Bacillus subtilis
To be Published
|
|
7U3A
 
 | | Structure of the Streptomyces venezuelae GlgX-c-di-GMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Glycogen debranching enzyme GlgX | | Authors: | Schumacher, M.A. | | Deposit date: | 2022-02-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Allosteric regulation of glycogen breakdown by the second messenger cyclic di-GMP.
Nat Commun, 13, 2022
|
|
8DYR
 
 | | BRD4-D1 in complex with BET inhibitor | | Descriptor: | (4P,6P)-4-[2-(cyclopropylmethoxy)-5-(methanesulfonyl)phenyl]-6-[1-(2-fluoroethyl)-1H-1,2,3-triazol-4-yl]-2-methylisoquinolin-1(2H)-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Gorman, M.A, Fitzgerald, C.G.D, White, J.M, Parker, M.W. | | Deposit date: | 2022-08-04 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Bromodomain and extraterminal protein-targeted probe enables tumour visualisation in vivo using positron emission tomography.
Chem.Commun.(Camb.), 59, 2023
|
|
8E17
 
 | | BRD4-D1 in complex with BET inhibitor | | Descriptor: | (4P,6M)-6-[1-(2-fluoroethyl)-1H-1,2,3-triazol-4-yl]-4-[5-(methanesulfonyl)-2-methoxyphenyl]-2-methylisoquinolin-1(2H)-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Gorman, M.A, Fitzgerald, C.G.D, White, J.M, Parker, M.W. | | Deposit date: | 2022-08-09 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Bromodomain and extraterminal protein-targeted probe enables tumour visualisation in vivo using positron emission tomography.
Chem.Commun.(Camb.), 59, 2023
|
|
8E3W
 
 | | BRD4-D1 in complex with BET inhibitor | | Descriptor: | (4P)-4-[2-(cyclopropylmethoxy)-5-(methanesulfonyl)phenyl]-2-methylisoquinolin-1(2H)-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Gorman, M.A, Fitzgerald, C.G.D, White, J.M, Parker, M.W. | | Deposit date: | 2022-08-17 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Bromodomain and extraterminal protein-targeted probe enables tumour visualisation in vivo using positron emission tomography.
Chem.Commun.(Camb.), 59, 2023
|
|
8G5I
 
 | | Cryo-EM structure of the Mismatch Sensing Complex (I) of Human Mitochondrial DNA Polymerase Gamma | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Nayak, A.R, Buchel, G, Herbine, K.H, Sarfallah, A, Sokolova, V.O, Zamudio-Ochoa, A, Temiakov, D. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural basis for DNA proofreading.
Nat Commun, 14, 2023
|
|
8G5J
 
 | | Cryo-EM structure of the Mismatch Uncoupling Complex (II) of Human Mitochondrial DNA Polymerase Gamma | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Nayak, A.R, Buchel, G, Herbine, K.H, Sarfallah, A, Sokolova, V.O, Zamudio-Ochoa, A, Temiakov, D. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structural basis for DNA proofreading.
Nat Commun, 14, 2023
|
|
