6ONG
 
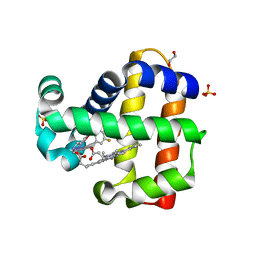 | | Dehaloperoxidate B in complex with substrate 4-F-cresol | | Descriptor: | 1,2-ETHANEDIOL, 4-fluoro-2-methylphenol, Dehaloperoxidase B, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, Malewschik, T, McGuire, A. | | Deposit date: | 2019-04-22 | | Release date: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The multifunctional globin dehaloperoxidase strikes again: Simultaneous peroxidase and peroxygenase mechanisms in the oxidation of EPA pollutants.
Arch.Biochem.Biophys., 673, 2019
|
|
7QBA
 
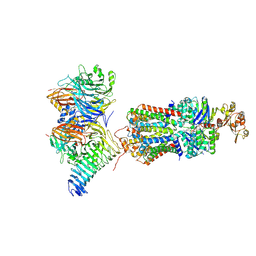 | | CryoEM structure of the ABC transporter NosDFY complexed with nitrous oxide reductase NosZ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Zipfel, S, Mueller, C, Topitsch, A, Lutz, M, Zhang, L, Einsle, O. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
1FCM
 
 | |
6UNX
 
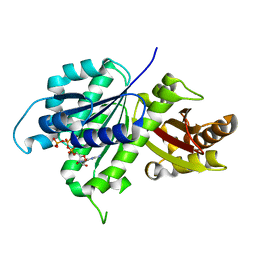 | | Structure of E. coli FtsZ(L178E)-GTP complex | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-10-13 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution crystal structures of Escherichia coli FtsZ bound to GDP and GTP.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7Q6L
 
 | |
7YVS
 
 | | Complex structure of Clostridioides difficile binary toxin unfolded CDTa-bound CDTb-pore (short). | | Descriptor: | ADP-ribosylating binary toxin binding subunit CdtB, ADP-ribosylating binary toxin enzymatic subunit CdtA, CALCIUM ION | | Authors: | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | Deposit date: | 2022-08-19 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
6OOU
 
 | | Crystal structure of HIV-1 Protease NL4-3 L89V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, NL4-3 PROTEASE, SULFATE ION | | Authors: | Henes, M, Kosovrasti, K, Lockbaum, G.J, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A, Whitfield, T.W. | | Deposit date: | 2019-04-23 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.127 Å) | | Cite: | Molecular Determinants of Epistasis in HIV-1 Protease: Elucidating the Interdependence of L89V and L90M Mutations in Resistance.
Biochemistry, 58, 2019
|
|
7YVQ
 
 | | Complex structure of Clostridioides difficile binary toxin folded CDTa-bound CDTb-pore (short). | | Descriptor: | ADP-ribosylating binary toxin binding subunit CdtB, ADP-ribosylating binary toxin enzymatic subunit CdtA, CALCIUM ION | | Authors: | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | Deposit date: | 2022-08-19 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
6Y0B
 
 | | Crystal structure of the cAMP-dependent protein kinase A cocrystallized with quinazolin-4-amine and PKI (5-24) | | Descriptor: | DIMETHYL SULFOXIDE, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha, ... | | Authors: | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2020-02-07 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Fragment based drug design - Small chemical changes of fragments effecting big changes in binding
To Be Published
|
|
2KYC
 
 | |
6GBO
 
 | | Crystal Structure of the oligomerization domain of Vp35 from Ebola virus | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Zinzula, L, Nagy, I, Orsini, M, Weyher-Stingl, E, Baumeister, W, Bracher, A. | | Deposit date: | 2018-04-16 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Ebola and Reston Virus VP35 Oligomerization Domains and Comparative Biophysical Characterization in All Ebolavirus Species.
Structure, 27, 2019
|
|
6OQU
 
 | | E. coli ATP synthase State 1d | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
7QC6
 
 | | HisF_C9A_L50H_I52H mutant (apo) from T. maritima | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Imidazole glycerol phosphate synthase subunit HisF | | Authors: | Beaumet, M, Dose, A, Braeuer, A, Mahy, J, Ghattas, W, Groll, M, Hess, C. | | Deposit date: | 2021-11-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An artificial metalloprotein with metal-adaptive coordination sites and Ni-dependent quercetinase activity.
J.Inorg.Biochem., 235, 2022
|
|
7L73
 
 | | Crystal structure of the first bromodomain (BD1) of human BRDT bound to ERK5-IN-1 | | Descriptor: | 11-cyclopentyl-2-({2-ethoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain testis-specific protein | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2020-12-25 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7QC7
 
 | | HisF-C9A-D11E-V33A_L50H_I52H mutant (apo) from T. maritima | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Imidazole glycerol phosphate synthase subunit HisF | | Authors: | Beaumet, M, Dose, A, Braeuer, A, Mahy, J, Ghattas, W, Groll, M, Hess, C. | | Deposit date: | 2021-11-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An artificial metalloprotein with metal-adaptive coordination sites and Ni-dependent quercetinase activity.
J.Inorg.Biochem., 235, 2022
|
|
6OPY
 
 | | HIV-1 Protease NL4-3 I13V, G16E, V32I, L33F, K45I, M46I, A71V, L76V, V82F, I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3 | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
8F0A
 
 | | Client-bound structure of a DegP trimer within a 12mer cage | | Descriptor: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | Authors: | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | Deposit date: | 2022-11-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
5M0P
 
 | | Crystal structure of cytochrome P450 OleT F79A in complex with arachidonic acid | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, Terminal olefin-forming fatty acid decarboxylase, ... | | Authors: | Tee, K.L, Munro, A, Matthews, S, Leys, D, Levy, C. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Catalytic Determinants of Alkene Production by the Cytochrome P450 Peroxygenase OleTJE.
J. Biol. Chem., 292, 2017
|
|
5QPV
 
 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000416a | | Descriptor: | (2~{S})-~{N}-(4-aminocarbonylphenyl)oxolane-2-carboxamide, Farnesyl diphosphate synthase, SULFATE ION, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
7QC8
 
 | | HisF-C9A-D11E-V33A_L50H_I52H mutant in complex with Zn(II) from T. maritima | | Descriptor: | Imidazole glycerol phosphate synthase subunit HisF, SULFATE ION, ZINC ION | | Authors: | Beaumet, M, Dose, A, Braeuer, A, Mahy, J, Ghattas, W, Groll, M, Hess, C. | | Deposit date: | 2021-11-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An artificial metalloprotein with metal-adaptive coordination sites and Ni-dependent quercetinase activity.
J.Inorg.Biochem., 235, 2022
|
|
7QC9
 
 | | HisF-C9A-D11E-V33A_L50H_I52H mutant in complex with Ni(II) from T. maritima | | Descriptor: | 1,2-ETHANEDIOL, Imidazole glycerol phosphate synthase subunit HisF, NICKEL (II) ION, ... | | Authors: | Beaumet, M, Dose, A, Braeuer, A, Mahy, J, Ghattas, W, Groll, M, Hess, C. | | Deposit date: | 2021-11-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An artificial metalloprotein with metal-adaptive coordination sites and Ni-dependent quercetinase activity.
J.Inorg.Biochem., 235, 2022
|
|
8QZC
 
 | |
2KPT
 
 | | Solution NMR structure of the N-terminal domain of cg2496 protein from Corynebacterium glutamicum. Northeast Structural Genomics Consortium Target CgR26A | | Descriptor: | Putative secreted protein | | Authors: | Eletsky, A, Sathyamoorthy, B, Sukumaran, D.K, Wang, D, Buchwald, W.A, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the N-terminal domain of cg2496 protein from Corynebacterium glutamicum
To be Published
|
|
5M1H
 
 | |
7UXG
 
 | | Crystal structure of putative serine protease YdgD from Escherichia coli | | Descriptor: | Serine protease | | Authors: | Stogios, P.J, Michalska, K, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-05-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structure of putative serine protease YdgD from Escherichia coli
To Be Published
|
|
