7Z2P
 
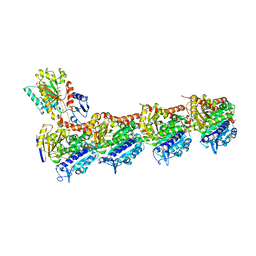 | | Tubulin-nocodazole complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Prota, A.E, Muehlethaler, T, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2022-02-28 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel fragment-derived colchicine-site binders as microtubule-destabilizing agents.
Eur.J.Med.Chem., 241, 2022
|
|
6EY5
 
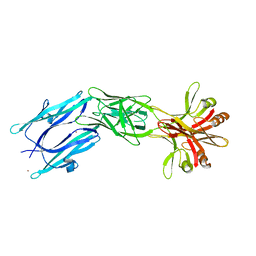 | | C-terminal part (residues 224-515) of PorM | | Descriptor: | T9SS component cytoplasmic membrane protein PorM, ZINC ION | | Authors: | Leone, P, Cambillau, C, Roussel, A. | | Deposit date: | 2017-11-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Type IX secretion system PorM and gliding machinery GldM form arches spanning the periplasmic space.
Nat Commun, 9, 2018
|
|
7Z2N
 
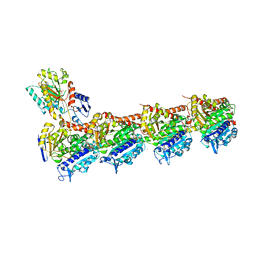 | | Tubulin-18-complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Muehlethaler, T, Prota, A.E, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2022-02-28 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel fragment-derived colchicine-site binders as microtubule-destabilizing agents.
Eur.J.Med.Chem., 241, 2022
|
|
8F1T
 
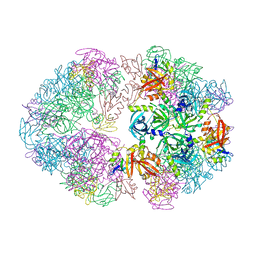 | | Structure of an 18mer DegP cage bound to the client protein hTRF1 | | Descriptor: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | Authors: | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | Deposit date: | 2022-11-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (12.1 Å) | | Cite: | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
7Z73
 
 | | Crystal structure of p63 tetramerization domain in complex with darpin 8F1 | | Descriptor: | Darpin 8F1, Isoform 2 of Tumor protein 63 | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
5MV9
 
 | | Structure of human Myosin 7a C-terminal MyTH4-FERM domain in complex with harmonin-a PDZ3 domain | | Descriptor: | ACETATE ION, Unconventional myosin-VIIa, cDNA FLJ51329, ... | | Authors: | Yu, I, Sourigues, Y, Titus, M.A, Houdusse, A. | | Deposit date: | 2017-01-16 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Myosin 7 and its adaptors link cadherins to actin.
Nat Commun, 8, 2017
|
|
6PXO
 
 | |
7AH1
 
 | | L19 diabody fragment from immunocytokine L19-IL2 | | Descriptor: | Anti-(ED-B) scFV | | Authors: | Ongaro, T, Guarino, S.R, Scietti, L, Palamini, M, Wulhfard, S, Villa, A, Neri, D, Forneris, F. | | Deposit date: | 2020-09-23 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inference of molecular structure for characterization and improvement of clinical grade immunocytokines.
J.Struct.Biol., 213, 2021
|
|
7NNG
 
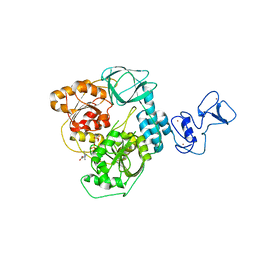 | | Crystal structure of the SARS-CoV-2 helicase in complex with Z2327226104 | | Descriptor: | 1-(2-methylphenyl)-1,2,3-triazole-4-carboxylic acid, PHOSPHATE ION, SARS-CoV-2 helicase NSP13, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Bountra, C, Gileadi, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
4R7W
 
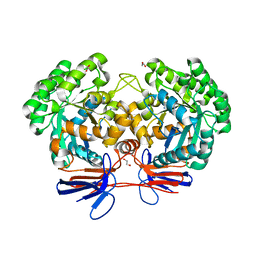 | | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with phosphonocytosine | | Descriptor: | (2R)-2-amino-2,5-dihydro-1,5,2-diazaphosphinin-6(1H)-one 2-oxide, 1,2-ETHANEDIOL, Cytosine deaminase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hitchcock, D.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with phosphonocytosine
To be Published
|
|
7NGC
 
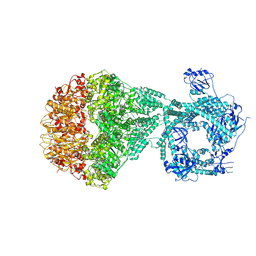 | | P2a-state of wild type human mitochondrial LONP1 protease with bound substrate protein and in presence of ATPgS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial, ... | | Authors: | Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T, Abrahams, J.P. | | Deposit date: | 2021-02-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
8B9O
 
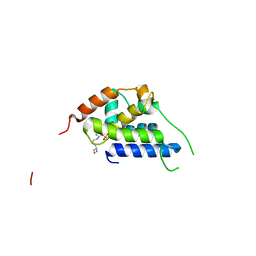 | |
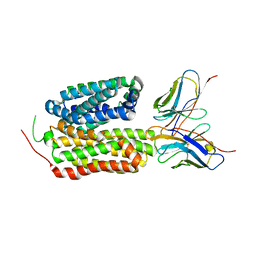
8HPJ
 
 | |
6YNO
 
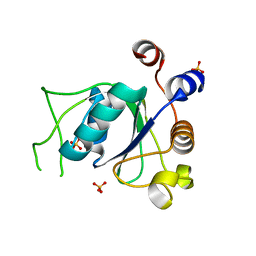 | | Crystal structure of YTHDC1 with compound DHU_DC1_139 | | Descriptor: | 6-[[methyl-[(1-phenylpyrazol-3-yl)methyl]amino]methyl]-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
7B1R
 
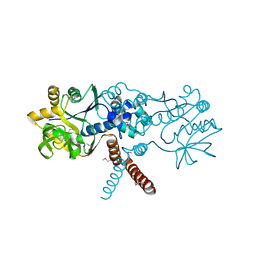 | |
5G57
 
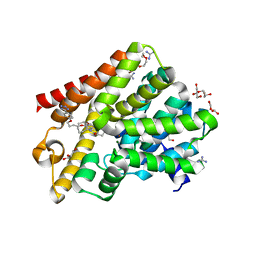 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-001 | | Descriptor: | (4~{a}~{S},8~{a}~{R})-2-cycloheptyl-4-[4-methoxy-3-[4-[4-(1~{H}-1,2,3,4-tetrazol-5-yl)phenoxy]butoxy]phenyl]-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-one, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2016-05-22 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
4RD8
 
 | | The crystal structure of a functionally-unknown protein from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | Uncharacterized protein | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-18 | | Release date: | 2014-10-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The crystal structure of a functionally-unknown protein from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To be Published
|
|
6VKX
 
 | | Crystal structure of the carbohydrate-binding domain VP8* of human P[8] rotavirus strain BM13851 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Outer capsid protein VP4, TETRAETHYLENE GLYCOL | | Authors: | Xu, S, McGinnis, K.R, Jiang, X, Kennedy, M.A. | | Deposit date: | 2020-01-22 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of P[II] rotavirus evolution and host ranges under selection of histo-blood group antigens.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5MYF
 
 | | Convergent evolution involving dimeric and trimeric dUTPases in signalling. | | Descriptor: | dUTPase from DI S. aureus phage | | Authors: | Donderis, J, Bowring, J, Maiques, E, Ciges-Tomas, J.R, Alite, C, Mehmedov, I, Tormo-Mas, M.A, Penades, J.R, Marina, A. | | Deposit date: | 2017-01-26 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Convergent evolution involving dimeric and trimeric dUTPases in pathogenicity island mobilization.
PLoS Pathog., 13, 2017
|
|
8BBY
 
 | |
4R10
 
 | | A conserved phosphorylation switch controls the interaction between cadherin and beta-catenin in vitro and in vivo | | Descriptor: | 1,2-ETHANEDIOL, Cadherin-related hmr-1, Protein humpback-2, ... | | Authors: | Choi, H.-J, Loveless, T, Lynch, A, Bang, I, Hardin, J, Weis, W.I. | | Deposit date: | 2014-08-03 | | Release date: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Conserved Phosphorylation Switch Controls the Interaction between Cadherin and beta-Catenin In Vitro and In Vivo
Dev.Cell, 33, 2015
|
|
4FAX
 
 | |
6YG6
 
 | | Crystal structure of MKK7 (MAP2K7) covalently bound with type-II inhibitor TL10-105 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 7, ~{N}-[4-[(4-ethylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl]-4-methyl-3-[2-[[(3~{S})-1-propanoylpyrrolidin-3-yl]amino]pyrimidin-4-yl]oxy-benzamide | | Authors: | Chaikuad, A, Tan, L, Wang, J, Liang, Y, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Catalytic Domain Plasticity of MKK7 Reveals Structural Mechanisms of Allosteric Activation and Diverse Targeting Opportunities.
Cell Chem Biol, 27, 2020
|
|
6YHD
 
 | | tRNA-guanine Transglycosylase (TGT) labeled with 5-fluorotryptophan in co-crystallized complex with 6-amino-4-(2-((2R,3S,4R,5R)-3,4-dihydroxy-5-methoxytetrahydrofuran-2-yl)ethyl)-2-(methylamino)-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | CHLORIDE ION, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Nguyen, A, Heine, A, Klebe, G. | | Deposit date: | 2020-03-28 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Co-crystallization, nanoESI-MS and 19F NMR reveal dimer disturbing inhibitors and conformational changes at dimer contacts
To Be Published
|
|
5MTU
 
 | | Maltodextrin binding protein MalE1 from L. casei BL23 bound to alpha-cyclodextrin | | Descriptor: | Cyclohexakis-(1-4)-(alpha-D-glucopyranose), MalE1 | | Authors: | Homburg, C, Bommer, M, Wuttge, S, Hobe, C, Beck, S, Dobbek, H, Deutscher, J, Licht, A, Schneider, E. | | Deposit date: | 2017-01-10 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.999 Å) | | Cite: | Inducer exclusion in Firmicutes: insights into the regulation of a carbohydrate ATP binding cassette transporter from Lactobacillus casei BL23 by the signal transducing protein P-Ser46-HPr.
Mol. Microbiol., 105, 2017
|
|
