8QZ1
 
 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with a nanobody (Nb58) | | Descriptor: | Isoform B of Potassium channel subfamily K member 10, Nanobody 58, POTASSIUM ION | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Baronina, A, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.588 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
7ZB5
 
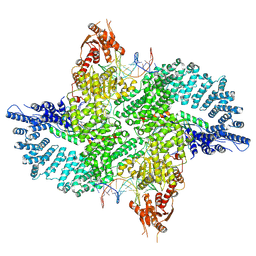 | | Mot1(1-1836):TBP:DNA - post-hydrolysis complex dimer | | Descriptor: | DNA (36-MER), Helicase-like protein, Putative tata-box binding protein | | Authors: | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | Deposit date: | 2022-03-23 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7BAR
 
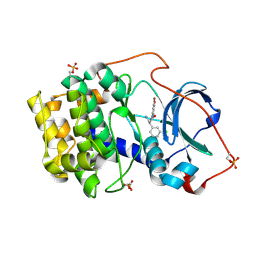 | |
8QQ7
 
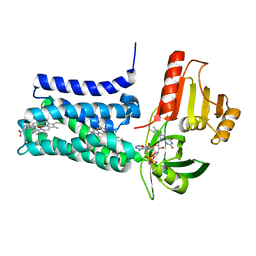 | | Structure of SpNOX: a Bacterial NADPH oxidase | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FAD-binding FR-type domain-containing protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Thepaut, M, Petit-Hartlein, I, Vermot, A, Chaptal, V, Humm, A.S, Dupeux, F, Marquez, J.A, Smith, S, Fieschi, F. | | Deposit date: | 2023-10-04 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | X-ray structure and enzymatic study of a bacterial NADPH oxidase highlight the activation mechanism of eukaryotic NOX.
Elife, 13, 2024
|
|
6YDK
 
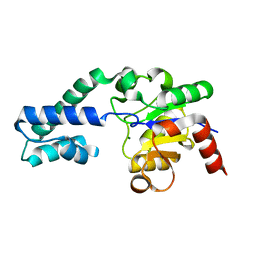 | | Substrate-free P146A variant of beta-phosphoglucomutase from Lactococcus lactis | | Descriptor: | Beta-phosphoglucomutase, MAGNESIUM ION | | Authors: | Wood, H.P, Cruz-Navarrete, F.A, Baxter, N.J, Trevitt, C.R, Robertson, A.J, Dix, S.R, Hounslow, A.M, Cliff, M.J, Waltho, J.P. | | Deposit date: | 2020-03-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Allomorphy as a mechanism of post-translational control of enzyme activity.
Nat Commun, 11, 2020
|
|
8R9O
 
 | | A soakable crystal form of human CDK7 in complex with AMP-PNP | | Descriptor: | Cyclin-dependent kinase 7, ~{N}-[(1~{S})-2-(dimethylamino)-1-phenyl-ethyl]-6,6-dimethyl-3-[[4-(propanoylamino)phenyl]carbonylamino]-1,4-dihydropyrrolo[3,4-c]pyrazole-5-carboxamide | | Authors: | Mukherjee, M, Cleasby, A. | | Deposit date: | 2023-11-30 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Protein engineering enables a soakable crystal form of human CDK7 primed for high-throughput crystallography and structure-based drug design.
Structure, 2024
|
|
6UBY
 
 | | Isolated cofilin bound to an actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Huehn, A.R, Bibeau, J.P, Schramm, A.C, Cao, W, De La Cruz, E.M, Sindelar, C.V. | | Deposit date: | 2019-09-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structures of cofilin-induced structural changes reveal local and asymmetric perturbations of actin filaments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8QQ1
 
 | | SpNOX dehydrogenase domain, mutant F397W in complex with Flavin adenine dinucleotide (FAD) | | Descriptor: | BROMIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Oxidoreductase | | Authors: | Humm, A.S, Dupeux, F, Vermot, A, Petit-Harleim, I, Fieschi, F, Marquez, J.A. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | X-ray structure and enzymatic study of a bacterial NADPH oxidase highlight the activation mechanism of eukaryotic NOX.
Elife, 13, 2024
|
|
5NER
 
 | |
6OGP
 
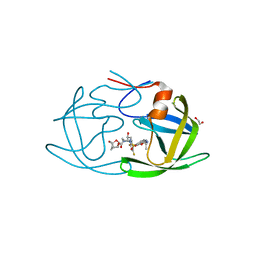 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-063 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-1-(3,5-difluorophenyl)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzoxazol-6-yl}sulfonyl)amino]butan-2-yl}carbamate, 1,2-ETHANEDIOL, Protease | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
6OGT
 
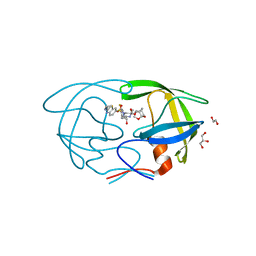 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P51) in complex with GRL-001 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
5NFG
 
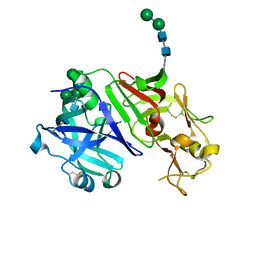 | | Structure of recombinant cardosin B from Cynara cardunculus | | Descriptor: | Procardosin-B,Procardosin-B, alpha-D-mannopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Pereira, P.J.B, Figueiredo, A.C, Manso, J.A, Almeida, C.M, Simoes, I. | | Deposit date: | 2017-03-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.375 Å) | | Cite: | Functional and structural characterization of synthetic cardosin B-derived rennet.
Appl. Microbiol. Biotechnol., 101, 2017
|
|
8QQ5
 
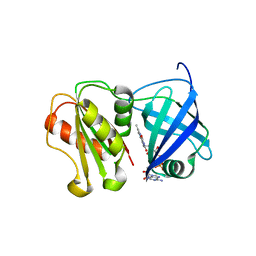 | | Structure of WT SpNox DH domain: a bacterial NADPH oxidase. | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Oxidoreductase | | Authors: | Thepaut, M, Petit-Hartlein, I, Vermot, A, Humm, A.S, Dupeux, F, Marquez, J.A, Smith, S, Fieschi, F. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure and enzymatic study of a bacterial NADPH oxidase highlight the activation mechanism of eukaryotic NOX.
Elife, 13, 2024
|
|
8DRL
 
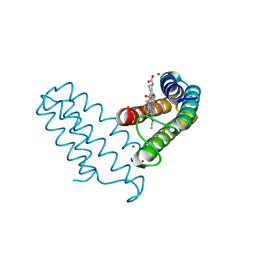 | |
7TJ4
 
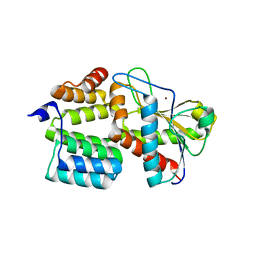 | | Structure of the S. aureus amidase LytH and activator ActH extracellular domains | | Descriptor: | ActH, LytH, ZINC ION | | Authors: | Page, J.E, Skiba, M.A, Kruse, A.C, Walker, S. | | Deposit date: | 2022-01-14 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Metal cofactor stabilization by a partner protein is a widespread strategy employed for amidase activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6GJR
 
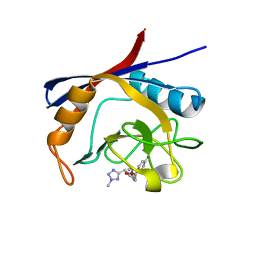 | | Cyclophilin A complexed with tri-vector ligand 9. | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, ethyl 2-[[(4-aminophenyl)methyl-[(2-methyl-1,2,3,4-tetrazol-5-yl)methyl]carbamoyl]amino]ethanoate | | Authors: | Georgiou, C, De Simone, A, Walkinshaw, M.D, Michel, J. | | Deposit date: | 2018-05-16 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A computationally designed binding mode flip leads to a novel class of potent tri-vector cyclophilin inhibitors.
Chem Sci, 10, 2019
|
|
6GJY
 
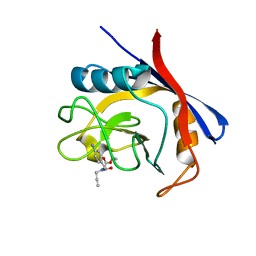 | | Cyclophilin A complexed with tri-vector ligand 5. | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, ethyl 2-[[(4-aminophenyl)methyl-prop-2-ynyl-carbamoyl]amino]ethanoate | | Authors: | Georgiou, C, De Simone, A, Walkinshaw, M.D, Michel, J. | | Deposit date: | 2018-05-17 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | A computationally designed binding mode flip leads to a novel class of potent tri-vector cyclophilin inhibitors.
Chem Sci, 10, 2019
|
|
5KVC
 
 | |
8DRF
 
 | | Zn(II)-bound B2 dimer (H60/H100/H104) | | Descriptor: | HEME C, Soluble cytochrome b562, ZINC ION | | Authors: | Choi, T.S, Tezcan, F.A. | | Deposit date: | 2022-07-20 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of a Flexible, Zn-Selective Protein Scaffold that Displays Anti-Irving-Williams Behavior.
J.Am.Chem.Soc., 144, 2022
|
|
7BBY
 
 | |
5KWH
 
 | | Crystal structure of CK2 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-07-18 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of CK2
To Be Published
|
|
6UKX
 
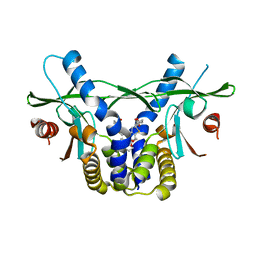 | | STING C-terminal Domain Complexed with Non-cyclic Dinucleotide Compound 11 | | Descriptor: | 4,4'-{propane-1,3-diylbis[oxy(5-methoxy-1-benzothiene-6,2-diyl)]}bis(4-oxobutanoic acid), fusion protein of Ubiquitin-like protein SMT3 and Stimulator of interferon protein c-terminal domain | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-10-06 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | An orally available non-nucleotide STING agonist with antitumor activity.
Science, 369, 2020
|
|
6UKZ
 
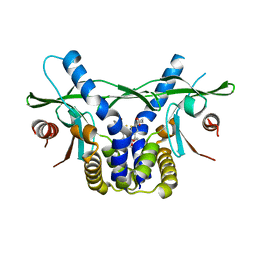 | | STING C-terminal Domain Complexed with Non-cyclic Dinucleotide Compound 6 | | Descriptor: | 4-[5-(2-{[2-(3-carboxypropanoyl)-4-fluoro-6-methoxy-1-benzothiophen-5-yl]oxy}ethoxy)-6-methoxy-1-benzothiophen-2-yl]-4-oxobutanoic acid, fusion protein of Ubiquitin-like protein SMT3 and Stimulator of interferon protein c-terminal domain | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-10-06 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | An orally available non-nucleotide STING agonist with antitumor activity.
Science, 369, 2020
|
|
8EAT
 
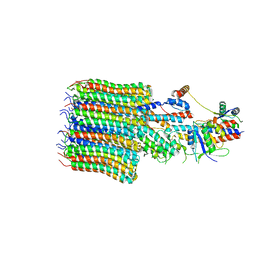 | | Yeast VO missing subunits a, e, and f in complex with Vma12-22p | | Descriptor: | V-type proton ATPase subunit F, V-type proton ATPase subunit c, V-type proton ATPase subunit c', ... | | Authors: | Wang, H, Bueler, S.A, Rubinstein, J.L. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of V-ATPase V O region assembly by Vma12p, 21p, and 22p.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5NI2
 
 | |
