4WAA
 
 | | Crystal structure of Nix LIR-fused human LC3B_2-119 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Suzuki, H, Ravichandran, A.C, Dobson, R.C.J, Novak, I, Wakatsuki, S. | | Deposit date: | 2014-08-29 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Phosphorylation of the mitochondrial autophagy receptor Nix enhances its interaction with LC3 proteins.
Sci Rep, 7, 2017
|
|
7NTM
 
 | |
4UXF
 
 | | Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fibre protein gp34, P21 native crystal | | Descriptor: | GLYCEROL, LARGE TAIL FIBER PROTEIN P34 | | Authors: | Granell, M, Alvira, S, Garcia-Doval, C, Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34.
Viruses, 9, 2017
|
|
4V0G
 
 | | JAK3 in complex with a covalent EGFR inhibitor | | Descriptor: | N-[3-(2-{3-amino-6-[1-(1-methylpiperidin-4-yl)-1H-pyrazol-4-yl]pyrazin-2-yl}-1H-benzimidazol-1-yl)phenyl]propanamide, TYROSINE-PROTEIN KINASE JAK3 | | Authors: | Debreczeni, J.E, Hennessy, E.J, Chuaquini, C, Ashton, S, Coclough, N, Cross, D.A.E, Eberlein, C, Gingipalli, L, Klinowska, T.C.M, Orme, J.P, Sha, L, Wu, X. | | Deposit date: | 2014-09-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Utilisation of Structure Based Design to Identify Novel, Irreversible Inhibitors of the Epidermal Growth Factor Receptor (Egfr) Harboring the Gatekeeper T790M Mutation
To be Published
|
|
6KSY
 
 | |
6KSG
 
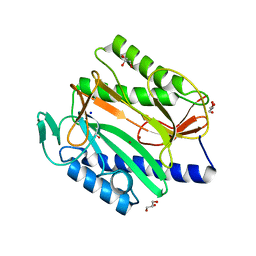 | | Vibrio cholerae Methionine Aminopeptidase in holo form | | Descriptor: | GLYCEROL, Methionine aminopeptidase, NICKEL (II) ION, ... | | Authors: | Pillalamarri, V, Addlagatta, A. | | Deposit date: | 2019-08-23 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Methionine aminopeptidases with short sequence inserts within the catalytic domain are differentially inhibited: Structural and biochemical studies of three proteins from Vibrio spp.
Eur.J.Med.Chem., 209, 2020
|
|
7P7D
 
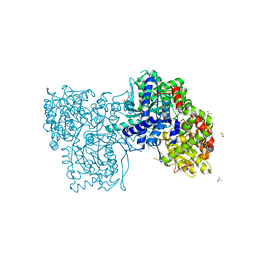 | | Rabbit muscle Glycogen Phosphorylase T state | | Descriptor: | DIMETHYL SULFOXIDE, Glycogen phosphorylase, muscle form | | Authors: | Stravodimos, G.A, Leonidas, D.D. | | Deposit date: | 2021-07-19 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Glycogen phosphorylase revisited: extending the resolution of the R- and T-state structures of the free enzyme and in complex with allosteric activators.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
6KRH
 
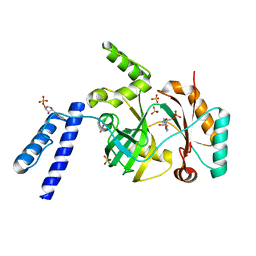 | | Structural basis for domain rotation during adenylation of active site K123 and fragment library screening against NAD+ -dependent DNA ligase from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase A, ... | | Authors: | Ramachandran, R, Shukla, A, Afsar, M. | | Deposit date: | 2019-08-21 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Salt bridges at the subdomain interfaces of the adenylation domain and active-site residues of Mycobacterium tuberculosis NAD + -dependent DNA ligase A (MtbLigA) are important for the initial steps of nick-sealing activity.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6F61
 
 | |
7P8K
 
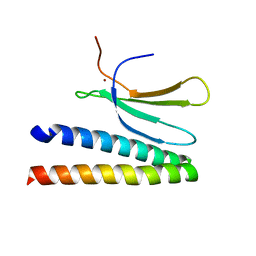 | | Crystal structure of in planta processed AvrRps4 in complex with the WRKY domain of RRS1 | | Descriptor: | Avirulence protein,Avirulence protein, Disease resistance protein RRS1, ZINC ION | | Authors: | Mukhi, N, Brown, H, Gorenkin, D, Ding, P, Bentham, A.R, Jones, J.D.G, Banfield, M.J. | | Deposit date: | 2021-07-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Perception of structurally distinct effectors by the integrated WRKY domain of a plant immune receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7QRZ
 
 | |
7QS2
 
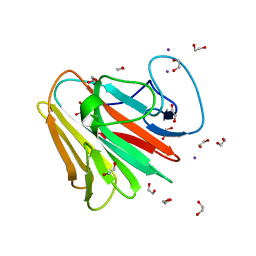 | |
7QS3
 
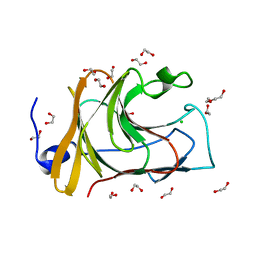 | |
7OW9
 
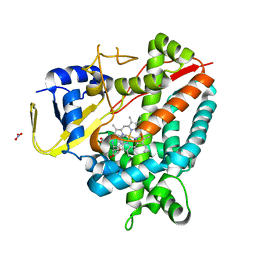 | | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with Cyclo-L-leucyl-L-leucine | | Descriptor: | (3S,6S)-3,6-bis(2-methylpropyl)piperazine-2,5-dione, CYPX, NITRATE ION, ... | | Authors: | Snee, M, Levy, C, Leys, D, Katariya, M, Munro, A.W. | | Deposit date: | 2021-06-17 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with Cyclo-L-leucyl-L-leucine
To Be Published
|
|
5PHM
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D in complex with N09455a | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, N-OXALYLGLYCINE, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5AMN
 
 | | The Discovery of 2-Substituted Phenol Quinazolines as Potent and Selective RET Kinase Inhibitors | | Descriptor: | 4-[3-HYDROXYANILINO]-6,7-DIMETHOXYQUINAZOLINE, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Newton, R, Bowler, K, Burns, E.M, Chapman, P, Fairweather, E, Fritzl, S, Goldberg, K, Hamilton, N.M, Holt, S.V, Hopkins, G.V, Jones, S.D, Jordan, A.M, Lyons, A, McDonald, N.Q, Maguire, L.A, Mould, D.P, Purkiss, A.G, Small, H.F, Stowell, A, Thomson, G.J, Waddell, I.D, Waszkowycz, B, Watson, A.J, Ogilvie, D.J. | | Deposit date: | 2015-03-11 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The discovery of 2-substituted phenol quinazolines as potent RET kinase inhibitors with improved KDR selectivity.
Eur J Med Chem, 112, 2016
|
|
7Q3R
 
 | |
6KO8
 
 | | Crystal structure of the Cholic acid bound RamR determined with XtaLAB Synergy | | Descriptor: | CHOLIC ACID, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
6FF1
 
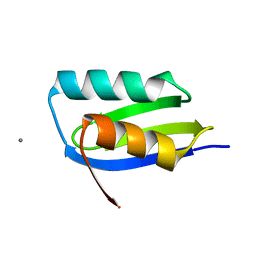 | | CsoZ metallochaperone | | Descriptor: | CALCIUM ION, CsoZ | | Authors: | Pelicioli-Riboldi, G, Basle, A, Waldron, K. | | Deposit date: | 2018-01-03 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The copper homeostasis system of Staphylococcus aureus:
Two putative metallochaperones with contrasting copper binding properties.
To Be Published
|
|
8Q4F
 
 | | Structure of arbekacin bound Escherichia coli 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 50S ribosomal protein L16, ... | | Authors: | Majumdar, S, Parajuli, N.P, Ge, X, Emmerich, A, Sanyal, S. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of arbekacin bound Escherichia coli 70S ribosome
To be published
|
|
7TVD
 
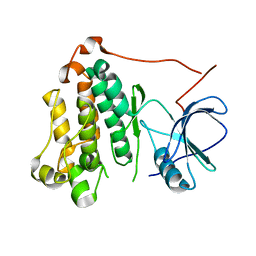 | |
4X5U
 
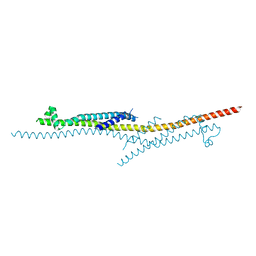 | | X-ray crystal structure of CagL at pH 4.2 | | Descriptor: | Cag pathogenicity island protein (Cag18) | | Authors: | Sundberg, E.J, Bonsor, D.A, Diederichs, K. | | Deposit date: | 2014-12-05 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Integrin Engagement by the Helical RGD Motif of the Helicobacter pylori CagL Protein Is Regulated by pH-induced Displacement of a Neighboring Helix.
J.Biol.Chem., 290, 2015
|
|
5C7K
 
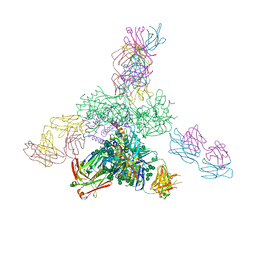 | | Crystal structure BG505 SOSIP gp140 HIV-1 Env trimer bound to broadly neutralizing antibodies PGT128 and 8ANC195 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab 8ANC195 heavy chain, ... | | Authors: | Kong, L, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2015-06-24 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.6019 Å) | | Cite: | Complete epitopes for vaccine design derived from a crystal structure of the broadly neutralizing antibodies PGT128 and 8ANC195 in complex with an HIV-1 Env trimer.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6L2C
 
 | | Crystal structure of Aspergillus fumigatus mitochondrial acetyl-CoA acetyltransferase in complex with CoA | | Descriptor: | Acetyl-CoA-acetyltransferase, putative, COENZYME A | | Authors: | Zhang, Y, Wei, W, Raimi, O.G, Ferenbach, A.T, Fang, W. | | Deposit date: | 2019-10-03 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Aspergillus fumigatus Mitochondrial Acetyl Coenzyme A Acetyltransferase as an Antifungal Target.
Appl.Environ.Microbiol., 86, 2020
|
|
5CU1
 
 | |
