6B4F
 
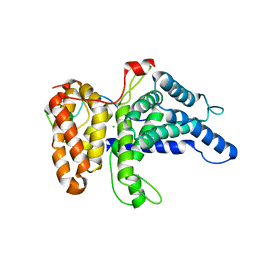 | | Crystal structure of human Gle1 CTD-Nup42 GBM complex | | Descriptor: | CHLORIDE ION, Nucleoporin GLE1, Nucleoporin like 2, ... | | Authors: | Lin, D.H, Correia, A.R, Cai, S.W, Huber, F.M, Jette, C.A, Hoelz, A. | | Deposit date: | 2017-09-26 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.811 Å) | | Cite: | Structural and functional analysis of mRNA export regulation by the nuclear pore complex.
Nat Commun, 9, 2018
|
|
5SZT
 
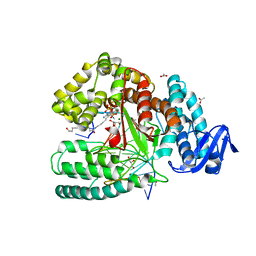 | | Crystal structure of the large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with 7-(N-(10-hydroxydecanoyl)-aminopentenyl)-7-deaza-2'-dATP | | Descriptor: | 7-(N-(10-hydroxydecanoyl)-aminopentenyl)-7-deaza-2'-dATP, ACETATE ION, DNA (5'-D(*AP*AP*AP*TP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Hottin, A, Betz, K, Marx, A. | | Deposit date: | 2016-08-15 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for the KlenTaq DNA Polymerase Catalysed Incorporation of Alkene- versus Alkyne-Modified Nucleotides.
Chemistry, 23, 2017
|
|
6L1X
 
 | | Quinol-dependent nitric oxide reductase (qNOR) from Neisseria meningitidis in the monomeric oxidized state with zinc complex. | | Descriptor: | CALCIUM ION, FE (III) ION, Nitric-oxide reductase, ... | | Authors: | Jamali, M.M.A, Antonyuk, S.V, Tosha, T, Muramoto, K, Hasnain, S.S, Shiro, Y. | | Deposit date: | 2019-10-01 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The active form of quinol-dependent nitric oxide reductase fromNeisseria meningitidisis a dimer.
Iucrj, 7, 2020
|
|
6VFM
 
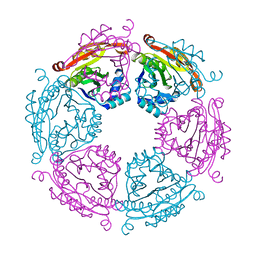 | | Crystal structure of SpeG allosteric polyamine acetyltransferase from Bacillus thuringiensis | | Descriptor: | Spermidine N1-acetyltransferase | | Authors: | Tsimbalyuk, S, Shornikov, A, Le, V.T.B, Kuhn, M.L, Forwood, J.K. | | Deposit date: | 2020-01-05 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | SpeG polyamine acetyltransferase enzyme from Bacillus thuringiensis forms a dodecameric structure and exhibits high catalytic efficiency.
J.Struct.Biol., 210, 2020
|
|
3LCO
 
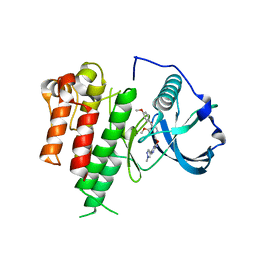 | | Inhibitor Bound to A DFG-Out structure of the Kinase Domain of CSF-1R | | Descriptor: | 3-({4-methoxy-5-[(4-methoxybenzyl)oxy]pyridin-2-yl}methoxy)-5-(1-methyl-1H-pyrazol-4-yl)pyrazin-2-amine, Macrophage colony-stimulating factor 1 receptor | | Authors: | Kamtekar, S, Day, J.E, Reitz, B.A, Mathis, K.J, Meyers, M.J. | | Deposit date: | 2010-01-11 | | Release date: | 2010-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure-based drug design enables conversion of a DFG-in binding CSF-1R kinase inhibitor to a DFG-out binding mode.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4RI5
 
 | |
4Y44
 
 | | Endothiapepsin in complex with fragment 164 | | Descriptor: | 4,6-dimethyl-2-{[3-(morpholin-4-yl)propyl]amino}pyridine-3-carbonitrile, ACETATE ION, Endothiapepsin, ... | | Authors: | Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
6IPU
 
 | |
6B8U
 
 | | Crystals Structure of B-Raf kinase domain in complex with an Imidazopyridinyl benzamide inhibitor | | Descriptor: | Serine/threonine-protein kinase B-raf, ~{N}-[3-(2-acetamidoimidazo[1,2-a]pyridin-6-yl)-4-methyl-phenyl]-3-(trifluoromethyl)benzamide | | Authors: | Appleton, B.A, Murray, J, Shafer, C.M. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Imidazo[1,2-a]pyridin-6-yl-benzamide analogs as potent RAF inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
3LEZ
 
 | |
3LOX
 
 | | HCV NS3-4a protease domain with a ketoamide inhibitor derivative of Boceprevir bound | | Descriptor: | (1R,2S,5S)-N-[(2S,3R)-4-amino-1-cyclobutyl-3-hydroxy-4-oxobutan-2-yl]-6,6-dimethyl-3-{3-methyl-N-[(1-methylcyclohexyl)c arbamoyl]-L-valyl}-3-azabicyclo[3.1.0]hexane-2-carboxamide, BETA-MERCAPTOETHANOL, HCV NS3 Protease, ... | | Authors: | Prongay, A.J. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The introduction of P4 substituted 1-methylcyclohexyl groups into Boceprevir: a change in direction in the search for a second generation HCV NS3 protease inhibitor.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5T2S
 
 | | Structure of the FHA1 domain of Rad53 bound simultaneously to the BRCT domain of Dbf4 and a phosphopeptide. | | Descriptor: | ASP-GLY-GLU-SER-TPO-ASP-GLU-ASP-ASP, DDK kinase regulatory subunit DBF4,Serine/threonine-protein kinase RAD53, GLYCEROL | | Authors: | Guarne, A, Almawi, A, Matthews, L. | | Deposit date: | 2016-08-24 | | Release date: | 2016-10-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 'AND' logic gates at work: Crystal structure of Rad53 bound to Dbf4 and Cdc7.
Sci Rep, 6, 2016
|
|
5G26
 
 | | Unveiling the Mechanism Behind the in-meso Crystallization of Membrane Proteins | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, INTIMIN | | Authors: | Zabara, A, Newman, J, Peat, T.S. | | Deposit date: | 2016-04-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The Nanoscience Behind the Art of in-Meso Crystallization of Membrane Proteins.
Nanoscale, 9, 2017
|
|
6R5I
 
 | |
3LG5
 
 | | F198A Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, ... | | Authors: | Aaron, J.A, Lin, X, Cane, D.E, Christianson, D.W. | | Deposit date: | 2010-01-19 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Structure of Epi-Isozizaene Synthase from Streptomyces coelicolor A3(2), a Platform for New Terpenoid Cyclization Templates
Biochemistry, 49, 2010
|
|
4YV3
 
 | |
6EVM
 
 | | Crystal structure of a Pro-9 complexed peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase | | Descriptor: | DIMETHYL SULFOXIDE, Pro-9, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
2X4O
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to HIV-1 envelope peptide env120-128 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, ENVELOPE GLYCOPROTEIN GP160, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
3LRH
 
 | |
5CW8
 
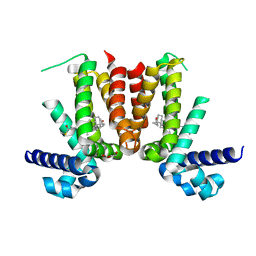 | | Crystal structure of Mycobacterium tuberculosis KstR in complex with 3-oxo-4-cholesten-26-oyl-CoA | | Descriptor: | HTH-type transcriptional repressor KstR, S-{1-[5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,7-dihydroxy-6,6-dimethyl-3-oxido-8,12 -dioxo-2,4-dioxa-9,13-diaza-3lambda~5~-phosphapentadecan-15-yl} (2S,6R)-6-[(8S,9S,10R,13R,14S,17R)-10,13-dimethyl-3-oxo-2,3,6,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta [a]phenanthren-17-yl]-2-methylheptanethioate (non-preferred name), TRIETHYLENE GLYCOL | | Authors: | Ho, N.A.T, Dawes, S, Kendall, S, Casabon, I, Crowe, A.M, Baker, E.N, Eltis, L.D, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-07-27 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure of the Transcriptional Repressor KstR in Complex with CoA Thioester Cholesterol Metabolites Sheds Light on the Regulation of Cholesterol Catabolism in Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
6BDS
 
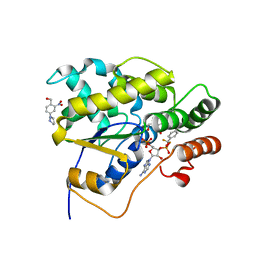 | | Schistosoma mansoni (Blood Fluke) Sulfotransferase/CIDD-0000204 (Compound 11f) Complex | | Descriptor: | (2-nitro-4-{[(3S)-1-{[4-(trifluoromethyl)phenyl]methyl}pyrrolidin-3-yl]amino}phenyl)methanol, ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase oxamniquine resistance protein | | Authors: | Taylor, A.B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Design, Synthesis, and Characterization of Novel Small Molecules as Broad Range Antischistosomal Agents.
ACS Med Chem Lett, 9, 2018
|
|
7JVF
 
 | |
5CXG
 
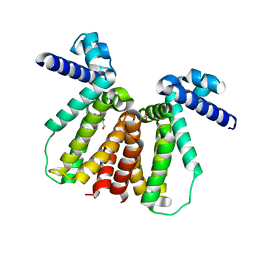 | | Crystal structure of Mycobacterium tuberculosis KstR in complex with PEG | | Descriptor: | DI(HYDROXYETHYL)ETHER, HTH-type transcriptional repressor KstR, TRIETHYLENE GLYCOL | | Authors: | Ho, N.A.T, Dawes, S, Kendall, S, Baker, E.N, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-07-28 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1001 Å) | | Cite: | The Structure of the Transcriptional Repressor KstR in Complex with CoA Thioester Cholesterol Metabolites Sheds Light on the Regulation of Cholesterol Catabolism in Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
5FIC
 
 | | Open form of murine Acid Sphingomyelinase in presence of lipid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Illes, K, Heinz, L.X, Superti-Furga, G, Nagar, B. | | Deposit date: | 2015-12-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of mammalian acid sphingomyelinase.
Nat Commun, 7, 2016
|
|
4RJ2
 
 | |
