6G38
 
 | | Crystal structure of haspin in complex with tubercidin | | Descriptor: | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Heroven, C, Chaikuad, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-24 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Halogen-Aromatic pi Interactions Modulate Inhibitor Residence Times.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7TKT
 
 | | SthK closed state, cAMP-bound in the presence of detergent | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Putative transcriptional regulator, ... | | Authors: | Rheinberger, J, Schmidpeter, P.A, Nimigean, C.M. | | Deposit date: | 2022-01-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Anionic lipids unlock the gates of select ion channels in the pacemaker family.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5UM1
 
 | | XFEL structure of influenza A M2 wild type TM domain at intermediate pH in the lipidic cubic phase at room temperature | | Descriptor: | CALCIUM ION, CHLORIDE ION, Matrix protein 2 | | Authors: | Thomaston, J.L, Woldeyes, R.A, Fraser, J.S, DeGrado, W.F. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | XFEL structures of the influenza M2 proton channel: Room temperature water networks and insights into proton conduction.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8VD3
 
 | |
6BKG
 
 | | Human LigIV catalytic domain with bound DNA-adenylate intermediate in closed conformation | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Moon, A.F, Tumbale, P.P, Schellenberg, M.J, Williams, R.S, Williams, J.G, Kunkel, T.A, Pedersen, L.C, Bebenek, B. | | Deposit date: | 2017-11-08 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structures of DNA-bound human ligase IV catalytic core reveal insights into substrate binding and catalysis.
Nat Commun, 9, 2018
|
|
6MTS
 
 | | Crystal structure of VRC43.03 Fab | | Descriptor: | Antibody VRC43.03 Fab heavy chain, Antibody VRC43.03 Fab light chain | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.437 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
6TLE
 
 | | Human Eg5 motor domain mutant E344K | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, MAGNESIUM ION, ... | | Authors: | Garcia-Saez, I, Skoufias, D.A. | | Deposit date: | 2019-12-02 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Human Eg5 motor domain mutant E344K
To Be Published
|
|
6G3C
 
 | | Crystal Structure of JAK2-V617F pseudokinase domain in complex with Compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]amino]-5,7,7-trimethyl-8-(3-methylbutyl)pteridin-6-one, Tyrosine-protein kinase | | Authors: | Dekker, C, Hinniger, A. | | Deposit date: | 2018-03-24 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
Acs Chem.Biol., 14, 2019
|
|
6SYB
 
 | | Crystal structure of carbonic anhydrase 2 with (3aR,4S,9bS)-4-(2-chloro-4-hydroxyphenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide | | Descriptor: | (3~{a}~{R},4~{S},9~{b}~{S})-4-(2-chloranyl-4-oxidanyl-phenyl)-2,3,3~{a},4,5,9~{b}-hexahydro-1~{H}-cyclopenta[c]quinoline-8-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2019-09-27 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of carbonic anhydrase 2 with (3aR,4S,9bS)-4-(2-chloro-4-hydroxyphenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide
To Be Published
|
|
8VJ4
 
 | | X-ray Counterpart to the Neutron Structure of Peroxide-Soaked Trp161Phe MnSOD | | Descriptor: | HYDROGEN PEROXIDE, MANGANESE (II) ION, Superoxide dismutase [Mn], ... | | Authors: | Azadmanesh, J, Slobodnik, K, Struble, L.R, Lutz, W.E, Weiss, K.L, Myles, D.A.A, Kroll, T, Borgstahl, G.E.O. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Revealing the atomic and electronic mechanism of human manganese superoxide dismutase product inhibition.
Biorxiv, 2024
|
|
8VHW
 
 | | Neutron Structure of Peroxide-Soaked Trp161Phe MnSOD | | Descriptor: | HYDROGEN PEROXIDE, MANGANESE (II) ION, Superoxide dismutase [Mn], ... | | Authors: | Azadmanesh, J, Slobodnik, K, Struble, L.R, Lutz, W.E, Coates, L, Weiss, K.L, Myles, D.A.A, Kroll, T, Borgstahl, G.E.O. | | Deposit date: | 2024-01-02 | | Release date: | 2024-07-31 | | Method: | NEUTRON DIFFRACTION (2.3 Å) | | Cite: | Revealing the atomic and electronic mechanism of human manganese superoxide dismutase product inhibition.
Biorxiv, 2024
|
|
7TJ6
 
 | | SthK open state, cAMP-bound in the presence of POPA | | Descriptor: | (2R)-1-(hexadecanoyloxy)-3-(phosphonooxy)propan-2-yl (9Z)-octadec-9-enoate, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Putative transcriptional regulator, ... | | Authors: | Schmidpeter, P.A, Nimigean, C.M. | | Deposit date: | 2022-01-14 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Anionic lipids unlock the gates of select ion channels in the pacemaker family.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5DJH
 
 | | Structure of M. tuberculosis CysQ, a PAP phosphatase with AMP, PO4, and 3Mg bound | | Descriptor: | 3'-phosphoadenosine 5'-phosphate phosphatase, ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fisher, A.J, Erickson, A.I. | | Deposit date: | 2015-09-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Crystal Structures of Mycobacterium tuberculosis CysQ, with Substrate and Products Bound.
Biochemistry, 54, 2015
|
|
6FPA
 
 | | Crystal structure of anti-mTFP1 DARPin 1238_G01 in space group P212121 | | Descriptor: | DARPin 1238_G01 | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6T07
 
 | | Crystal structure of YTHDC1 with fragment 20 (DHU_DC1_134) | | Descriptor: | SULFATE ION, YTH domain-containing protein 1, ~{N}-[(2~{S})-pyrrolidin-2-yl]-1~{H}-1,2,4-triazol-5-amine | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
8VHY
 
 | | Neutron Structure of Reduced Trp161Phe MnSOD | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Azadmanesh, J, Slobodnik, K, Struble, L.R, Lutz, W.E, Coates, L, Weiss, K.L, Myles, D.A.A, Kroll, T, Borgstahl, G.E.O. | | Deposit date: | 2024-01-02 | | Release date: | 2024-07-31 | | Method: | NEUTRON DIFFRACTION (2.3 Å) | | Cite: | Revealing the atomic and electronic mechanism of human manganese superoxide dismutase product inhibition.
Biorxiv, 2024
|
|
5FJD
 
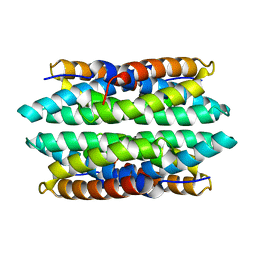 | | APO-CSP1 (COPPER STORAGE PROTEIN 1) FROM METHYLOSINUS TRICHOSPORIUM OB3B | | Descriptor: | COPPER STORAGE PROTEIN 1 | | Authors: | Vita, N, Platsaki, S, Basle, A, Allen, S.J, Paterson, N.G, Crombie, A.T, Murrell, J.C, Waldron, K.J, Dennison, C. | | Deposit date: | 2015-10-07 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Four-Helix Bundle Stores Copper for Methane Oxidation.
Nature, 525, 2015
|
|
6MUY
 
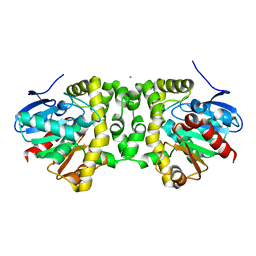 | | Fluoroacetate dehalogenase, room temperature structure solved by serial 3 degree oscillation crystallography | | Descriptor: | CALCIUM ION, Fluoroacetate dehalogenase | | Authors: | Finke, A.D, Wierman, J.L, Pare-Labrosse, O, Sarrachini, A, Besaw, J, Mehrabi, P, Gruner, S.M, Miller, R.J.D. | | Deposit date: | 2018-10-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fixed-target serial oscillation crystallography at room temperature.
IUCrJ, 6, 2019
|
|
8VJ0
 
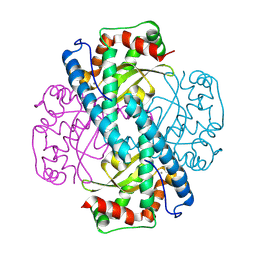 | | Neutron Structure of Oxidized Trp161Phe MnSOD | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Azadmanesh, J, Slobodnik, K, Struble, L.R, Lutz, W.E, Weiss, K.L, Myles, D.A.A, Kroll, T, Borgstahl, G.E.O. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-31 | | Method: | NEUTRON DIFFRACTION (2.3 Å) | | Cite: | Revealing the atomic and electronic mechanism of human manganese superoxide dismutase product inhibition.
Biorxiv, 2024
|
|
5DN3
 
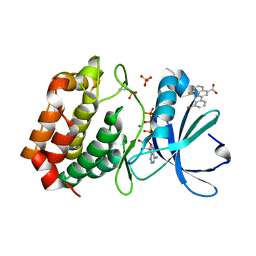 | | Aurora A in complex with ATP and AA35. | | Descriptor: | 2-(3-bromophenyl)-8-fluoroquinoline-4-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, ... | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G, Huggins, D, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A. | | Deposit date: | 2015-09-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
7TO0
 
 | | Cryo-EM structure of RIG-I in complex with OHdsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, OHdsRNA, ZINC ION | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
6HI3
 
 | | The ATAD2 bromodomain in complex with compound 4 | | Descriptor: | 2-azanyl-~{N}-(4-ethanoyl-1,3-thiazol-2-yl)-2-methyl-propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2018-08-29 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
8VD1
 
 | | Crystal Structure of KAI2 from Oryza sativa with MPD | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Probable esterase D14L | | Authors: | Gilio, A.K, Shabek, N, Guercio, A.M, Pawlak, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structural insights into rice KAI2 receptor provide functional implications for perception and signal transduction.
J.Biol.Chem., 300, 2024
|
|
6MVL
 
 | |
4FAR
 
 | | Structure of Oceanobacillus iheyensis group II intron in the presence of K+, Mg2+ and 5'-exon | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Group IIC intron, MAGNESIUM ION, ... | | Authors: | Marcia, M, Pyle, A.M. | | Deposit date: | 2012-05-22 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Visualizing Group II Intron Catalysis through the Stages of Splicing.
Cell(Cambridge,Mass.), 151, 2012
|
|
