6ZIJ
 
 | | Crystal Structure of Two-Domain Laccase mutant R240H from Streptomyces griseoflavus | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kolyadenko, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The role of positive charged residue in the proton-transfer mechanism of two-domain laccase from Streptomyces griseoflavus Ac-993.
J.Biomol.Struct.Dyn., 40, 2022
|
|
6XCB
 
 | |
5MG2
 
 | | Crystal structure of the second bromodomain of human TAF1 in complex with BAY-299 chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 6-(3-oxidanylpropyl)-2-(1,3,6-trimethyl-2-oxidanylidene-benzimidazol-5-yl)benzo[de]isoquinoline-1,3-dione, Transcription initiation factor TFIID subunit 1 | | Authors: | Tallant, C, Bouche, L, Holton, S.J, Fedorov, O, Siejka, P, Picaud, S, Krojer, T, Srikannathasan, V, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hartung, I.V, Haendler, B, Muller, S, Huber, K.V.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Benzoisoquinolinediones as Potent and Selective Inhibitors of BRPF2 and TAF1/TAF1L Bromodomains.
J. Med. Chem., 60, 2017
|
|
5WE0
 
 | | Structural Basis for Shelterin Bridge Assembly | | Descriptor: | DNA-binding protein rap1, Protection of telomeres protein poz1, Protection of telomeres protein tpz1, ... | | Authors: | Kim, J.-K, Liu, J, Hu, X, Yu, C, Roskamp, K, Sankaran, B, Huang, L, Komives, E.-A, Qiao, F. | | Deposit date: | 2017-07-06 | | Release date: | 2017-12-20 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Shelterin Bridge Assembly.
Mol. Cell, 68, 2017
|
|
5FPX
 
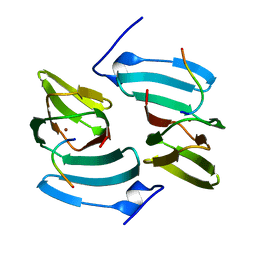 | | The structure of KdgF from Yersinia enterocolitica. | | Descriptor: | NICKEL (II) ION, PECTIN DEGRADATION PROTEIN, PEPTIDE | | Authors: | Hobbs, J.K, Lee, S.M, Robb, M, Hof, F, Barr, C, Abe, K.T, Hehemann, J.H, McLean, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2015-12-03 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Kdgf, the Missing Link in the Microbial Metabolism of Uronate Sugars from Pectin and Alginate.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5MHO
 
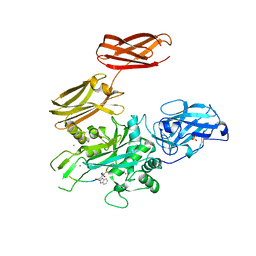 | |
1NQT
 
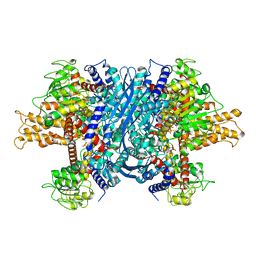 | | Crystal structure of bovine Glutamate dehydrogenase-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamate dehydrogenase 1 | | Authors: | Banerjee, S, Schmidt, T, Fang, J, Stanley, C.A, Smith, T.J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation
Biochemistry, 42, 2003
|
|
8QAQ
 
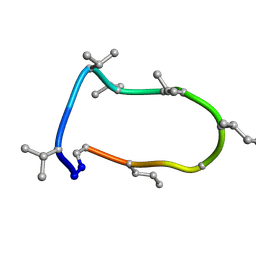 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability. Conformation 1 of omphalotin A in apolar solvents. | | Descriptor: | TRP-MVA-ILE-MVA-MVA-SAR-MVA-IML-SAR-VAL-IML-SAR | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
6RVU
 
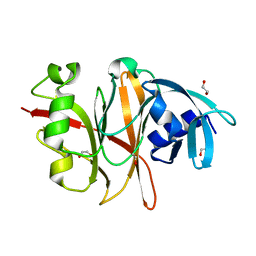 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) | | Descriptor: | 1,2-ETHANEDIOL, Lethal Factor 1 (BLF1) | | Authors: | Mobbs, G.W, Aziz, A.A, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | Deposit date: | 2019-06-01 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
7TR8
 
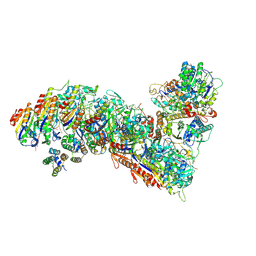 | | Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-10 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
7TR9
 
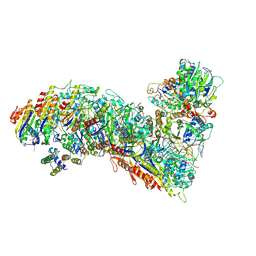 | | Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
6UAM
 
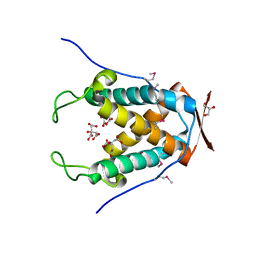 | |
8AWW
 
 | | Transthyretin conjugated with a tafamidis derivative | | Descriptor: | Transthyretin, ~{N}-(6-azanylhexyl)-2-[3,5-bis(chloranyl)phenyl]-1,3-benzoxazole-6-carboxamide | | Authors: | Cerofolini, L, Vasa, K, Bianconi, E, Salobehaj, M, Cappelli, G, Licciardi, G, Perez-Rafols, A, Padilla Cortes, L.D, Antonacci, S, Rizzo, D, Ravera, E, Calderone, V, Parigi, G, Luchinat, C, Macchiarulo, A, Menichetti, S, Fragai, M. | | Deposit date: | 2022-08-30 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Combining Solid-State NMR with Structural and Biophysical Techniques to Design Challenging Protein-Drug Conjugates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5MKM
 
 | |
6OBI
 
 | | Remarkable rigidity of the single alpha-helical domain of myosin-VI revealed by NMR spectroscopy | | Descriptor: | Myosin-VI | | Authors: | Barnes, A, Shen, Y, Ying, J, Takagi, Y, Torchia, D.A, Sellers, J, Bax, A. | | Deposit date: | 2019-03-20 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Remarkable Rigidity of the Single alpha-Helical Domain of Myosin-VI As Revealed by NMR Spectroscopy.
J.Am.Chem.Soc., 141, 2019
|
|
6YX9
 
 | | Cryogenic human adiponectin receptor 2 (ADIPOR2) at 2.4 A resolution determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 2, GLYCEROL, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6ROH
 
 | | Cryo-EM structure of the autoinhibited Drs2p-Cdc50p | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Timcenko, M, Lyons, J.A, Januliene, D, Ulstrup, J.J, Dieudonne, T, Montigny, C, Ash, M.R, Karlsen, J.L, Boesen, T, Kuhlbrandt, W, Lenoir, G, Moeller, A, Nissen, P. | | Deposit date: | 2019-05-13 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and autoregulation of a P4-ATPase lipid flippase.
Nature, 571, 2019
|
|
6IGR
 
 | | Crystal structure of S9 peptidase (S514A mutant in inactive state) from Deinococcus radiodurans R1 | | Descriptor: | Acyl-peptide hydrolase, putative, GLYCEROL | | Authors: | Yadav, P, Gaur, N.K, Goyal, V.D, Kumar, A, Makde, R.D. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
3OFG
 
 | | Structured Domain of Caenorhabditis elegans BMY-1 | | Descriptor: | Boca/mesd chaperone for ywtd beta-propeller-egf protein 1, CHLORIDE ION | | Authors: | Collins, M.N, Hendrickson, W.A. | | Deposit date: | 2010-08-14 | | Release date: | 2011-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.367 Å) | | Cite: | Structural Characterization of the Boca/Mesd Maturation Factors for LDL-Receptor-Type beta-Propeller Domains
Structure, 2011
|
|
6YXD
 
 | | Room temperature structure of human adiponectin receptor 2 (ADIPOR2) at 2.9 A resolution determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 2, GLYCEROL, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
7U8Z
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 (CD2) complexed with fluorinated peptoid inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-({N-[2-(benzylamino)-2-oxoethyl]-4-(dimethylamino)benzamido}methyl)-3-fluoro-N-hydroxybenzamide, ACETATE ION, ... | | Authors: | Watson, P.R, Cragin, A.D, Christianson, D.W. | | Deposit date: | 2022-03-09 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of Fluorinated Peptoid-Based Histone Deacetylase (HDAC) Inhibitors for Therapy-Resistant Acute Leukemia.
J.Med.Chem., 65, 2022
|
|
6AX2
 
 | |
8DRD
 
 | | Ni(II)-bound B2 dimer (H60/H100/H104) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, HEME C, NICKEL (II) ION, ... | | Authors: | Choi, T.S, Tezcan, F.A. | | Deposit date: | 2022-07-20 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Design of a Flexible, Zn-Selective Protein Scaffold that Displays Anti-Irving-Williams Behavior.
J.Am.Chem.Soc., 144, 2022
|
|
6BF2
 
 | |
3R15
 
 | | Structure Treponema Denticola Factor H Binding Protein | | Descriptor: | Factor H binding protein, THIOCYANATE ION | | Authors: | Miller, D.P, McDowell, J.V, Burgner, J, Heroux, A, Bell, J.K, Marconi, R.T. | | Deposit date: | 2011-03-09 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structure Treponema Denticola Factor H Binding Protein
To be Published
|
|
