8DU7
 
 | | Room-temperature serial synchrotron crystallography (SSX) structure of apo PTP1B | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Sharma, S, Keedy, D.A. | | Deposit date: | 2022-07-27 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Room-temperature serial synchrotron crystallography of the human phosphatase PTP1B.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8DTT
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV93-03 | | Descriptor: | COV93-03 heavy chain, COV93-03 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8QBP
 
 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability. NMR structure of Omphalotin A in methanol / water indoleOut conformation. | | Descriptor: | TRP-MVA-ILE-MVA-MVA-SAR-MVA-IML-SAR-VAL-IML-SAR | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
5MFB
 
 | | Designed armadillo repeat protein YIII(Dq)4CqI | | Descriptor: | YIII(Dq)4CqI | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
6ANM
 
 | |
1ZPV
 
 | | ACT domain protein from Streptococcus pneumoniae | | Descriptor: | ACT domain protein, POTASSIUM ION | | Authors: | Osipiuk, J, Hatzos, C, Abdullah, J, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-17 | | Release date: | 2005-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystal structure of ACT domain protein from Streptococcus pneumoniae
To be Published
|
|
6LYC
 
 | | Crystal structure of the NOD SIRPa complex with D4-2 | | Descriptor: | ACETIC ACID, D4-2, SIRPa of the NOD mouse strain | | Authors: | Murata, Y, Matsuda, M, Nakagawa, A, Matozaki, T. | | Deposit date: | 2020-02-14 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Macrocyclic Peptide-Mediated Blockade of the CD47-SIRP alpha Interaction as a Potential Cancer Immunotherapy.
Cell Chem Biol, 27, 2020
|
|
6SCT
 
 | | Cryo-EM structure of the consensus triskelion hub of the clathrin coat complex | | Descriptor: | Clathrin heavy chain, Clathrin light chain | | Authors: | Morris, K.L, Cameron, A.D, Sessions, R, Smith, C.J. | | Deposit date: | 2019-07-25 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | Cryo-EM of multiple cage architectures reveals a universal mode of clathrin self-assembly.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4S3G
 
 | | Structure of the F249X mutant of Phosphatidylinositol-specific phospholipase C from Staphylococcus aureus | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, ACETATE ION | | Authors: | He, T, Gershenson, A, Eyles, S.J, Gao, J, Roberts, M.F. | | Deposit date: | 2015-01-26 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fluorinated Aromatic Amino Acids Distinguish Cation-pi Interactions from Membrane Insertion.
J.Biol.Chem., 290, 2015
|
|
4LLE
 
 | |
6GYJ
 
 | | Mouse thymidylate synthase crystal soaked in phenolphthalein | | Descriptor: | 3,3-bis(4-hydroxyphenyl)-2-benzofuran-1-one, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Maj, P, Jarmula, A, Wilk, P, Weiss, M.S, Rode, W. | | Deposit date: | 2018-06-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thymidylate synthase fragment screening
To be published
|
|
7RCC
 
 | |
6MCB
 
 | |
6SFI
 
 | | Crystal structure of p38 alpha in complex with compound 75 (MCP33) | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]-2-[[[1-(2-methylphenyl)pyrazol-4-yl]carbonylamino]methyl]-1,3-thiazole-5-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6ZP0
 
 | | Structure of SARS-CoV-2 Spike Protein Trimer (single Arg S1/S2 cleavage site) in Closed State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2VKC
 
 | | Solution structure of the B3BP Smr domain | | Descriptor: | NEDD4-BINDING PROTEIN 2 | | Authors: | Diercks, T, Ab, E, Daniels, M.A, deJong, R.N, Besseling, R, Kaptein, R, Folkers, G.E. | | Deposit date: | 2007-12-18 | | Release date: | 2008-10-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Characterization of the DNA- Binding Activity of the B3BP-Smr Domain.
J.Mol.Biol., 383, 2008
|
|
5H40
 
 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with sophorose | | Descriptor: | CALCIUM ION, GLYCEROL, Uncharacterized protein, ... | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
4Y5J
 
 | | Drosophila melanogaster Mini spindles TOG3 | | Descriptor: | MINI SPINDLES TOG3 | | Authors: | Slep, K.C, Howard, A.E. | | Deposit date: | 2015-02-11 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Drosophila melanogaster Mini Spindles TOG3 Utilizes Unique Structural Elements to Promote Domain Stability and Maintain a TOG1- and TOG2-like Tubulin-binding Surface.
J.Biol.Chem., 290, 2015
|
|
6SDH
 
 | | Human Carbonic Anhydrase II in complex with an inhibitor soaked at a concentration of 5 mM | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Gloeckner, S, Haake, M, Heine, A, Klebe, G. | | Deposit date: | 2019-07-27 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Human Carbonic Anhydrase II in complex with an inhibitor soaked at a concentration of 5 mM
To Be Published
|
|
6ODC
 
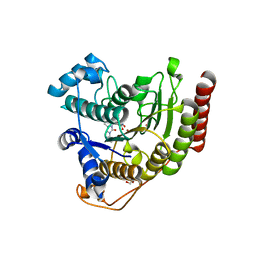 | | Crystal structure of HDAC8 in complex with compound 30 | | Descriptor: | (2E)-3-[2-(3-cyclopentyl-5,5-dimethyl-2-oxoimidazolidin-1-yl)phenyl]-N-hydroxyprop-2-enamide, 1,2-ETHANEDIOL, Histone deacetylase 8, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
6OJ2
 
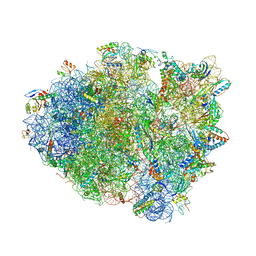 | | Crystal structure of tRNA^ Ala(GGC) bound to the near-cognate 70S A-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Nguyen, H.A, Sunita, S, Dunham, C.M. | | Deposit date: | 2019-04-10 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Disruption of evolutionarily correlated tRNA elements impairs accurate decoding.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6H1P
 
 | |
6Y1D
 
 | |
3M2Z
 
 | |
8DYN
 
 | |
