8GVB
 
 | | The complex between public TCR TD08 and HLA-A24 bound to HIV-1 Nef138-8 peptide | | Descriptor: | 8-mer peptide from Protein Nef, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Gao, G.F, Shi, Y, Ma, K, Chai, Y, Guan, J, Qi, J, Tan, S, Dong, T, Iwamoto, A, Kawana-Tachikawa, A. | | Deposit date: | 2022-09-14 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular Basis for the Recognition of HIV Nef138-8 Epitope by a Pair of Human Public T Cell Receptors.
J Immunol., 209, 2022
|
|
6SLD
 
 | | Structure of the Phosphatidylcholine Binding Mutant of Yeast Sec14 Homolog Sfh1 (S175I,T177I) in Complex with Phosphatidylinositol | | Descriptor: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CRAL-TRIO domain-containing protein YKL091C | | Authors: | Berger, J, Fitz, M, Johnen, P, Shanmugaratnam, S, Hocker, B, Stiel, A.C, Schaaf, G. | | Deposit date: | 2019-08-19 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Phosphatidylcholine Binding Mutant of Yeast Sec14 Homolog Sfh1 (S175I,T177I) in Complex with Phosphatidylinositol
To Be Published
|
|
8BGK
 
 | |
8DMH
 
 | | Lymphocytic choriomeningitis virus glycoprotein in complex with neutralizing antibody M28 | | Descriptor: | 18.5C-M28 Fab Heavy Chain, 18.5C-M28 Fab Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Moon-Walker, A, Hastie, K.M, Zyla, D.S, Saphire, E.O. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for antibody-mediated neutralization of lymphocytic choriomeningitis virus.
Cell Chem Biol, 30, 2023
|
|
6GZF
 
 | | Xi Class GST from Natrialba magadii | | Descriptor: | Glutathione S-transferase, SULFATE ION | | Authors: | Di Matteo, A, Federici, L, Masulli, M, Carletti, E, Cassidy, J, Paradisi, F, Di Ilio, C, Allocati, N. | | Deposit date: | 2018-07-04 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.614 Å) | | Cite: | Structural Characterization of the Xi Class Glutathione Transferase From the Haloalkaliphilic ArchaeonNatrialba magadii.
Front Microbiol, 10, 2019
|
|
6Y9N
 
 | | Crystal structure of Whirlin PDZ3_C-ter in complex with Myosin 15a C-terminal PDZ binding motif peptide | | Descriptor: | Unconventional myosin-XV, Whirlin | | Authors: | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | Deposit date: | 2020-03-10 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
6VDO
 
 | |
6Y9O
 
 | | Crystal structure of Whirlin PDZ3_C-ter in complex with CASK internal PDZ binding motif peptide | | Descriptor: | Peripheral plasma membrane protein CASK, Whirlin | | Authors: | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | Deposit date: | 2020-03-10 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.632 Å) | | Cite: | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
8DMI
 
 | | Lymphocytic choriomeningitis virus glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein G1, ... | | Authors: | Moon-Walker, A, Hastie, K.M, Zyla, D.S, Saphire, E.O. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis for antibody-mediated neutralization of lymphocytic choriomeningitis virus.
Cell Chem Biol, 30, 2023
|
|
8GVM
 
 | | The structure of azide-bound cytochrome C oxidase determined using the crystals exposed to 20 mm azide solution for 4 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimada, A. | | Deposit date: | 2022-09-15 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity
J. Biol. Chem., 293, 2018
|
|
5N07
 
 | |
8DV4
 
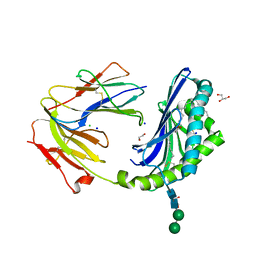 | | Crystal structure of the BC8B TCR-CD1b-PI complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Farquhar, R, Rossjohn, J, Shahine, A. | | Deposit date: | 2022-07-28 | | Release date: | 2023-04-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | alpha beta T-cell receptor recognition of self-phosphatidylinositol presented by CD1b.
J.Biol.Chem., 299, 2023
|
|
5N1I
 
 | |
6P8K
 
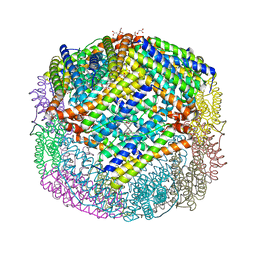 | | Escherichia coli Bacterioferritin Substituted with Zinc Protoporphyrin IX | | Descriptor: | Bacterioferritin, MALONATE ION, PROTOPORPHYRIN IX CONTAINING ZN, ... | | Authors: | Taylor, A.B, Cioloboc, D, Kurtz, D.M. | | Deposit date: | 2019-06-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a Zinc Porphyrin-Substituted Bacterioferritin and Photophysical Properties of Iron Reduction.
Biochemistry, 59, 2020
|
|
4QQN
 
 | | Protein arginine methyltransferase 3 in complex with compound MTV044246 | | Descriptor: | 1-{2-[1-(aminomethyl)cyclohexyl]ethyl}-3-isoquinolin-6-ylurea, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dong, A, Dobrovetsky, E, Tempel, W, He, H, Zhao, K, Smil, D, Landon, M, Luo, X, Chen, Z, Dai, M, Yu, Z, Lin, Y, Zhang, H, Zhao, K, Schapira, M, Brown, P.J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of Potent and Selective Allosteric Inhibitors of Protein Arginine Methyltransferase 3 (PRMT3).
J. Med. Chem., 61, 2018
|
|
8Z2G
 
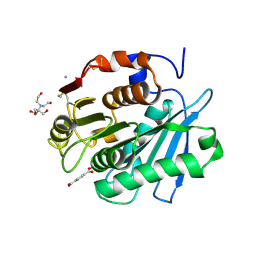 | | MHET bound form of PET-degrading cutinase mutant Cut190*SS_S176A | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-hydroxyethyloxycarbonyl)benzoic acid, AMMONIUM ION, ... | | Authors: | Numoto, N, Kondo, F, Bekker, G.J, Liao, Z, Yamashita, M, Iida, A, Ito, N, Kamiya, N, Oda, M. | | Deposit date: | 2024-04-12 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural dynamics of the Ca 2+ -regulated cutinase towards structure-based improvement of PET degradation activity.
Int.J.Biol.Macromol., 281, 2024
|
|
7N3S
 
 | |
8Z2H
 
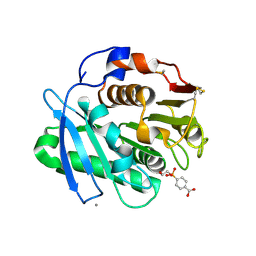 | | Substrate analog a010 bound form of PET-degrading cutinase mutant Cut190**SS_S176A | | Descriptor: | 4-[2-hydroxyethyloxy(oxidanyl)phosphoryl]benzoic acid, Alpha/beta hydrolase family protein, CALCIUM ION | | Authors: | Numoto, N, Kondo, F, Bekker, G.J, Liao, Z, Yamashita, M, Iida, A, Ito, N, Kamiya, N, Oda, M. | | Deposit date: | 2024-04-12 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural dynamics of the Ca 2+ -regulated cutinase towards structure-based improvement of PET degradation activity.
Int.J.Biol.Macromol., 281, 2024
|
|
4YPC
 
 | |
8Z2I
 
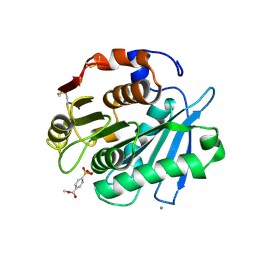 | | Substrate analog a011 bound form of PET-degrading cutinase mutant Cut190**SS_S176A | | Descriptor: | 2-hydroxyethyloxy-(4-methoxycarbonylphenyl)phosphinic acid, Alpha/beta hydrolase family protein, CALCIUM ION | | Authors: | Numoto, N, Kondo, F, Bekker, G.J, Liao, Z, Yamashita, M, Iida, A, Ito, N, Kamiya, N, Oda, M. | | Deposit date: | 2024-04-12 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural dynamics of the Ca 2+ -regulated cutinase towards structure-based improvement of PET degradation activity.
Int.J.Biol.Macromol., 281, 2024
|
|
5N49
 
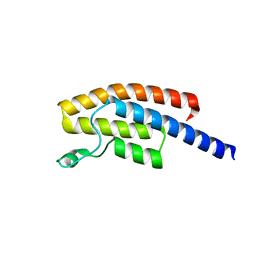 | | BRPF2 in complex with Compound 7 | | Descriptor: | 2-(1,3,6-trimethyl-2-oxidanylidene-benzimidazol-5-yl)benzo[de]isoquinoline-1,3-dione, Bromodomain-containing protein 1 | | Authors: | Bouche, L, Christ, C.D, Siegel, S, Fernandez-Montalvan, A.E, Holton, S.J, Fedorov, O, ter Laak, A, Sugawara, T, Stoeckigt, D, Tallant, C, Bennett, J, Monteiro, O, Saez, L.D, Siejka, P, Meier, J, Puetter, V, Weiske, J, Mueller, S, Huber, K.V.M, Hartung, I.V, Haendler, B. | | Deposit date: | 2017-02-10 | | Release date: | 2017-05-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Benzoisoquinolinediones as Potent and Selective Inhibitors of BRPF2 and TAF1/TAF1L Bromodomains.
J. Med. Chem., 60, 2017
|
|
1DSK
 
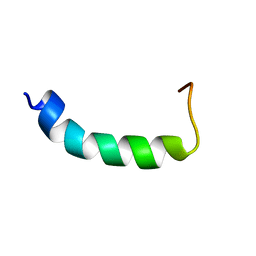 | | NMR SOLUTION STRUCTURE OF VPR59_86, 20 STRUCTURES | | Descriptor: | VPR PROTEIN | | Authors: | Yao, S, Torres, A.M, Azad, A.A, Macreadie, I.G, Norton, R.S. | | Deposit date: | 1997-10-23 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of peptides from HIV-1 Vpr protein that cause membrane permeabilization and growth arrest.
J. Pept. Sci., 4, 1998
|
|
7N3R
 
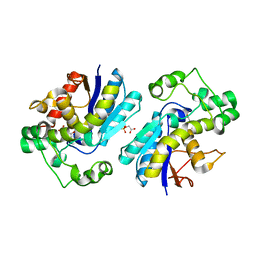 | |
8E20
 
 | |
7QUZ
 
 | | Crystal structure of the SeMet octameric C-terminal Big_2-CBM56 domains from Paenibacillus illinoisensis (Bacillus circulans IAM1165) beta-1,3-glucanase H | | Descriptor: | Beta-1,3-glucanase bglH, CHLORIDE ION, GLYCEROL | | Authors: | Najmudin, S, Venditto, I, Fontes, C.M.G.A, Bule, P. | | Deposit date: | 2022-01-19 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.156 Å) | | Cite: | Structural and biochemical characterization of C-terminal Big_2-CBM56 domains of Bacillus circulans IAM1165 beta-1,3-glucanase H and Paenibacillus sp CBM56
To be published
|
|
