7MYH
 
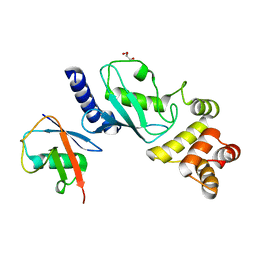 | | Ubiquitin variant UbV.k.2 in complex with Ube2k | | Descriptor: | GLYCEROL, Ubiquitin variant UbV.k.2, Ubiquitin-conjugating enzyme E2 K | | Authors: | Middleton, A.J, Day, C.L, Teyra, J, Sidhu, S.S. | | Deposit date: | 2021-05-21 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Identification of Ubiquitin Variants That Inhibit the E2 Ubiquitin Conjugating Enzyme, Ube2k.
Acs Chem.Biol., 16, 2021
|
|
8BS1
 
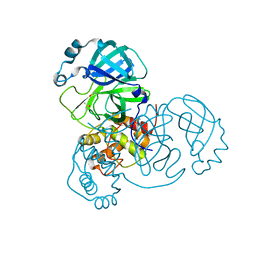 | | Room-temperature structure of SARS-CoV-2 Main protease at atmospheric pressure | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Lieske, J, Saouane, S, Guenther, S, Reinke, P.Y.A, Rahmani Mashhour, A, Meents, A. | | Deposit date: | 2022-11-24 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High-pressure macromolecular crystallography to explore the conformational space of proteins
To Be Published
|
|
7Z0N
 
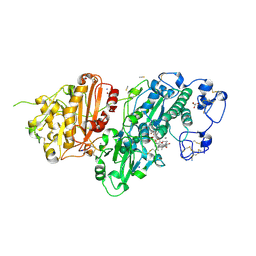 | | Structure-Based Design of a Novel Class of Autotaxin Inhibitors Based on Endogenous Allosteric Modulators | | Descriptor: | CALCIUM ION, GLYCEROL, IODIDE ION, ... | | Authors: | Salgado-Polo, F, Clark, J.M, Macdonald, S.J.F, Barrett, T.N, Perrakis, A, Jamieson, A. | | Deposit date: | 2022-02-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of a Novel Class of Autotaxin Inhibitors Based on Endogenous Allosteric Modulators.
J.Med.Chem., 65, 2022
|
|
7PM5
 
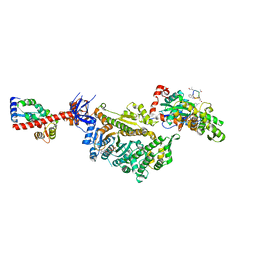 | | Cryo-EM structure of the actomyosin-V complex in the strong-ADP state (central 1er) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-02 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
4XHF
 
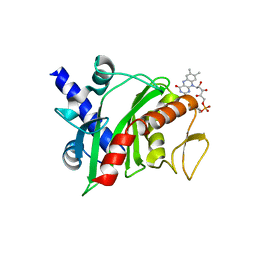 | | Crystal structure of Shewanella oneidensis NqrC | | Descriptor: | FLAVIN MONONUCLEOTIDE, Na-translocating NADH-quinone reductase subunit C NqrC, SODIUM ION | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2015-01-05 | | Release date: | 2015-12-16 | | Last modified: | 2016-03-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular insights into the enzymatic diversity of flavin-trafficking protein (Ftp; formerly ApbE) in flavoprotein biogenesis in the bacterial periplasm.
Microbiologyopen, 5, 2016
|
|
8BS2
 
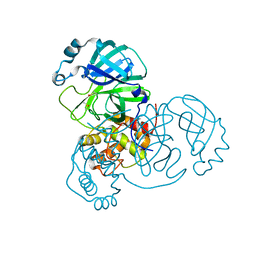 | | Room-temperature structure of SARS-CoV-2 Main protease at 104 MPa helium gas pressure in a sapphire capillary | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Lieske, J, Saouane, S, Guenther, S, Reinke, P.Y.A, Rahmani Mashhour, A, Meents, A. | | Deposit date: | 2022-11-24 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | High-pressure macromolecular crystallography to explore the conformational space of proteins
To Be Published
|
|
7PM9
 
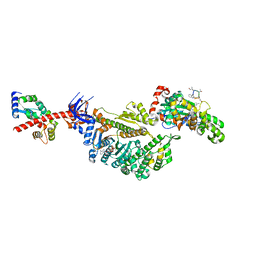 | | Cryo-EM structure of the actomyosin-V complex in the strong-ADP state (central 1er, class 4) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-02 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
5LF7
 
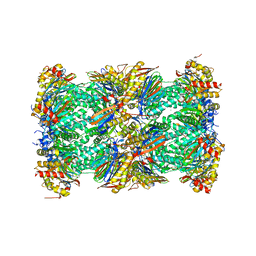 | | Human 20S proteasome complex with Ixazomib at 2.0 Angstrom | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Schrader, J, Henneberg, F, Mata, R, Tittmann, K, Schneider, T.R, Stark, H, Bourenkov, G, Chari, A. | | Deposit date: | 2016-06-30 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design.
Science, 353, 2016
|
|
6RJR
 
 | | Crystal structure of a Fungal Catalase at 1.9 Angstrom | | Descriptor: | CHLORIDE ION, Catalase, GLYCEROL, ... | | Authors: | Gomez, S, Navas-Yuste, S, Payne, A.M, Rivera, W, Lopez-Estepa, M, Brangbour, C, Fulla, D, Juanhuix, J, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2019-04-29 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Peroxisomal catalases from the yeasts Pichia pastoris and Kluyveromyces lactis as models for oxidative damage in higher eukaryotes.
Free Radic. Biol. Med., 141, 2019
|
|
7RU2
 
 | | SARS-CoV-2-6P-Mut7 S protein (asymmetric) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
6ENH
 
 | |
6UJC
 
 | | Integrin alpha-v beta-8 in complex with the Fabs C6-RGD3 and 11D12v2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C6-RGD3 heavy chain Fab, ... | | Authors: | Campbell, M.G, Cormier, A, Cheng, Y, Nishimura, S.L. | | Deposit date: | 2019-10-02 | | Release date: | 2020-02-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Cryo-EM Reveals Integrin-Mediated TGF-beta Activation without Release from Latent TGF-beta.
Cell, 180, 2020
|
|
4XBD
 
 | | 1.45A resolution structure of Norovirus 3CL protease complex with a covalently bound dipeptidyl inhibitor (1R,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid (Orthorhombic P Form) | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Weerawarna, P.M, Uy, R.A.Z, Damalanka, V.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.-O. | | Deposit date: | 2014-12-16 | | Release date: | 2015-03-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Guided Design and Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease. Structure-Activity Relationships and Biochemical, X-ray Crystallographic, Cell-Based, and In Vivo Studies.
J.Med.Chem., 58, 2015
|
|
7PMD
 
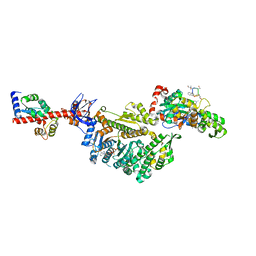 | | Cryo-EM structure of the actomyosin-V complex in the post-rigor transition state (AppNHp, central 1er) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-02 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
6U9A
 
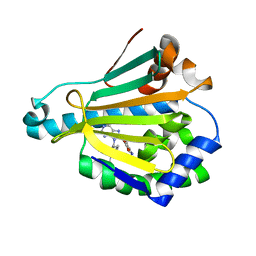 | | Hsp90a NTD K58R bound reversibly to sulfonyl fluoride 5 | | Descriptor: | 3-{[(3S)-3-({6-amino-8-[(6-iodo-2H-1,3-benzodioxol-5-yl)sulfanyl]-9H-purin-9-yl}methyl)piperidin-1-yl]methyl}benzene-1-sulfonyl fluoride, Heat shock protein HSP 90-alpha | | Authors: | Cuesta, A, Wan, X, Taunton, J. | | Deposit date: | 2019-09-07 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Ligand Conformational Bias Drives Enantioselective Modification of a Surface-Exposed Lysine on Hsp90.
J.Am.Chem.Soc., 142, 2020
|
|
5LHN
 
 | | The catalytic domain of murine urokinase-type plasminogen activator in complex with the allosteric inhibitory nanobody Nb7 | | Descriptor: | 1,2-ETHANEDIOL, Camelid-Derived Antibody Fragment Nb7, SULFATE ION, ... | | Authors: | Kromann-Hansen, T, Lange, E.L, Sorensen, H.P, Ghassabeh, G.H, Huang, M, Jensen, J.K, Muyldermans, S, Declerck, P, Andreasen, P.A. | | Deposit date: | 2016-07-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of a novel conformational equilibrium in urokinase-type plasminogen activator.
Sci Rep, 7, 2017
|
|
6ZTH
 
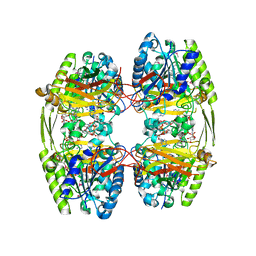 | |
6S1I
 
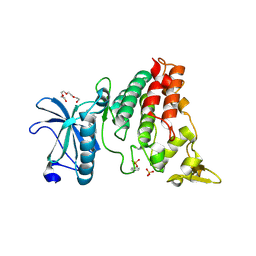 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Mining Public Domain Data to Develop Selective DYRK1A Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
7Z1Z
 
 | | MVV strand transfer complex (STC) intasome in complex with LEDGF/p75 at 3.5 A resolution | | Descriptor: | DNA (37-MER), DNA (5'-D(*GP*CP*TP*GP*CP*GP*AP*GP*AP*TP*CP*CP*GP*CP*TP*CP*CP*GP*GP*TP*G)-3'), DNA (5'-D(P*TP*TP*GP*AP*TP*TP*AP*GP*GP*GP*TP*G)-3'), ... | | Authors: | Pye, V.E, Ballandras-Colas, A, Cherepanov, P. | | Deposit date: | 2022-02-25 | | Release date: | 2022-05-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Multivalent interactions essential for lentiviral integrase function.
Nat Commun, 13, 2022
|
|
6OCR
 
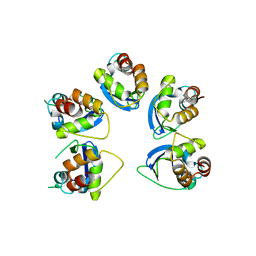 | | Crystal structure of human KCTD16 T1 domain | | Descriptor: | BTB/POZ domain-containing protein KCTD16 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6ES4
 
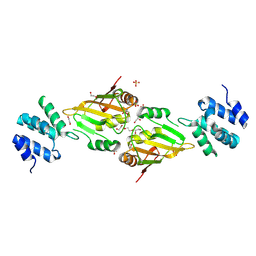 | | A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Syncrip, ... | | Authors: | Hobor, F, Dallmann, A, Ball, N.J, Cicchini, C, Battistelli, C, Ogrodowicz, R.W, Christodoulou, E, Martin, S.R, Castello, A, Tripodi, M, Taylor, I.A, Ramos, A. | | Deposit date: | 2017-10-19 | | Release date: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets.
Nat Commun, 9, 2018
|
|
6ODY
 
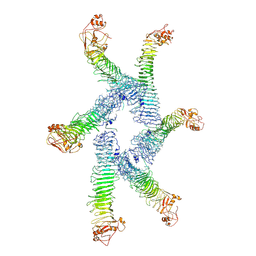 | |
7RWQ
 
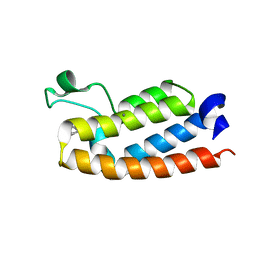 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one | | Descriptor: | 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7PM7
 
 | | Cryo-EM structure of the actomyosin-V complex in the strong-ADP state (central 1er, class 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-02 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
8EMZ
 
 | | Structure of GII.17 norovirus in complex with Nanobody 2 | | Descriptor: | 1,2-ETHANEDIOL, GII.17 P domain, Nanobody 2 | | Authors: | Kher, G, Sabin, C, Pancera, M, Koromyslova, A, Hansman, G. | | Deposit date: | 2022-09-28 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Direct Blockade of the Norovirus Histo-Blood Group Antigen Binding Pocket by Nanobodies.
J.Virol., 97, 2023
|
|
