5N1P
 
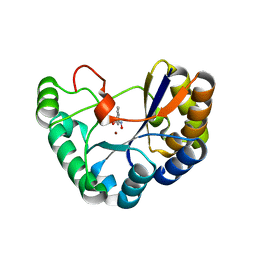 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with N-hydroxynaphthalene-1-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, Peptidoglycan N-acetylglucosamine deacetylase, SODIUM ION, ... | | Authors: | Giastas, P, Andreou, A, Eliopoulos, E.E. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
6NNZ
 
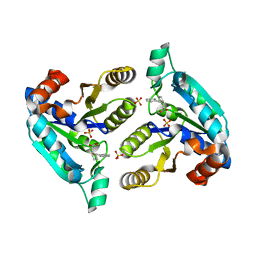 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 4 | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, SULFATE ION, [(1R,2S)-2-(benzenecarbonyl)cyclopentyl]acetic acid, ... | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-01-15 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 4
To Be Published
|
|
6XHL
 
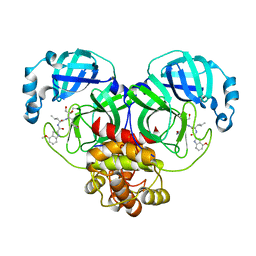 | | Covalent complex of SARS-CoV main protease with N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Descriptor: | 3C-like proteinase, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
6VR2
 
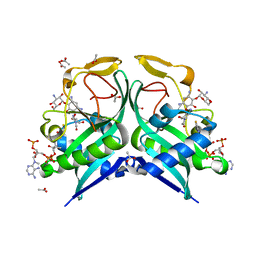 | | Aminoglycoside N-2'-Acetyltransferase-Ia [AAC(2')-Ia] in complex with acetylated-tobramycin and CoA | | Descriptor: | (1S,2S,3R,4S,6R)-3-{[2-(acetylamino)-6-amino-2,3,6-trideoxy-alpha-D-ribo-hexopyranosyl]oxy}-4,6-diamino-2-hydroxycycloh exyl 3-amino-3-deoxy-alpha-D-glucopyranoside, ACETATE ION, Aminoglycoside 2'-N-acetyltransferase, ... | | Authors: | Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2020-02-06 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and phylogenetic analyses of resistance to next-generation aminoglycosides conferred by AAC(2') enzymes.
Sci Rep, 11, 2021
|
|
5I2V
 
 | |
5N8D
 
 | |
5N3D
 
 | | cAMP-dependent Protein Kinase A from Cricetulus griseus in complex with fragment like molecule 4-(trifluoromethyl)benzenecarboximidamide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, [azanyl-[4-(trifluoromethyl)phenyl]methylidene]azanium, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Siefker, C, Heine, A, Klebe, G. | | Deposit date: | 2017-02-08 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Fragment Binding to Kinase Hinge: If Charge Distribution and Local pK a Shifts Mislead Popular Bioisosterism Concepts.
Angew.Chem.Int.Ed.Engl., 2020
|
|
6SUI
 
 | | AMICOUMACIN KINASE AMIN | | Descriptor: | PENTAETHYLENE GLYCOL, Phosphotransferase enzyme family protein | | Authors: | Bourenkov, G.P, Mokrushina, Y.A, Terekhov, S.S, Smirnov, I.V, Gabibov, A.G, Altman, S. | | Deposit date: | 2019-09-14 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A kinase bioscavenger provides antibiotic resistance by extremely tight substrate binding.
Sci Adv, 6, 2020
|
|
6NTC
 
 | | Crystal Structure of G12V HRas-GppNHp bound in complex with the engineered RBD variant 1 of CRAF Kinase protein | | Descriptor: | GLYCEROL, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Maisonneuve, P, Kurinov, I, Wiechmann, S, Ernst, A, Sicheri, F. | | Deposit date: | 2019-01-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Conformation-specific inhibitors of activated Ras GTPases reveal limited Ras dependency of patient-derived cancer organoids.
J.Biol.Chem., 295, 2020
|
|
7AU5
 
 | | Tubulin-noscapine-analogue-14e complex | | Descriptor: | (5~{R})-5-[(1~{S})-4,5-dimethoxy-1,3-dihydro-2-benzofuran-1-yl]-~{N}-ethyl-4-methoxy-7,8-dihydro-5~{H}-[1,3]dioxolo[4,5-g]isoquinoline-6-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Yong, C, Devine, S.M, Abel, A.-C, Muthiah, D, Gao, X, Callaghan, R, Capuano, B, Steinmetz, M.O, Prota, A.E, Scammels, P.J. | | Deposit date: | 2020-11-02 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 1,3-Benzodioxole-Modified Noscapine Analogues: Synthesis, Antiproliferative Activity, and Tubulin-Bound Structure.
Chemmedchem, 16, 2021
|
|
8AF8
 
 | | Room temperature SSX crystal structure of CTX-M-14 (5K dataset) | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Oberthuer, D, Perbandt, M, Prester, A, Rohde, H, Betzel, C, Yefanov, O. | | Deposit date: | 2022-07-15 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
5N4T
 
 | | VIM-2 metallo-beta-lactamase in complex with ((S)-3-mercapto-2-methylpropanoyl)-L-tryptophan (Compound 4) | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-2-[[(2~{S})-2-methyl-3-sulfanyl-propanoyl]amino]propanoic acid, BENZOIC ACID, Beta-lactamase VIM-2, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2017-02-11 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystallographic analyses of isoquinoline complexes reveal a new mode of metallo-beta-lactamase inhibition.
Chem. Commun. (Camb.), 53, 2017
|
|
7RCA
 
 | | Crystal structure of Aro2p chorismate synthase from Candida lusitaniae | | Descriptor: | CHLORIDE ION, Chorismate synthase, SULFATE ION | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-07 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of Aro2p chorismate synthase from Candida lusitaniae
To Be Published
|
|
6QJI
 
 | |
6NVM
 
 | |
7BK2
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 44 | | Descriptor: | 4-amino-7-methyl-2-({5-methyl-1-[(3R)-oxolan-3-yl]-1H-pyrazol-4-yl}amino)-6-[(2R)-2-methylpyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-15 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
5IV1
 
 | | Solution Structure of DNA Dodecamer with 8-oxoguanine at 4th Position | | Descriptor: | DNA (5'-D(*CP*GP*CP*(8OG)P*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Miears, H.L, Gruber, D.R, Hoppins, J.J, Kiryutin, A.S, Kasymov, R.D, Yurkovskaya, A.V, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 8-Oxoguanine Affects DNA Backbone Conformation in the EcoRI Recognition Site and Inhibits Its Cleavage by the Enzyme.
Plos One, 11, 2016
|
|
8F9O
 
 | | Dog sialic acid esterase (SIAE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sialic acid acetylesterase, ... | | Authors: | Ide, D, Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2022-11-24 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Dog sialic acid esterase (SIAE)
To Be Published
|
|
6NXA
 
 | | ECAII(D90T,K162T) MUTANT AT PH 7 | | Descriptor: | ACETIC ACID, GLYCEROL, L-asparaginase 2 | | Authors: | Lubkowski, J, Wlodawer, A. | | Deposit date: | 2019-02-08 | | Release date: | 2019-08-07 | | Last modified: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Opportunistic complexes of E. coli L-asparaginases with citrate anions.
Sci Rep, 9, 2019
|
|
8VEO
 
 | | Crystal structure of PRMT5:MEP50 in complex with MTA | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2023-12-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP -Deleted Cancers.
J.Med.Chem., 67, 2024
|
|
7BCX
 
 | | The adduct of NAMI-A with Hen Egg White Lysozyme at 8 hours. | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, IMIDAZOLE, ... | | Authors: | Chiniadis, L, Giastas, P, Bratsos, I, Papakyriakou, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Insights into the Protein Ruthenation Mechanism by Antimetastatic Metallodrugs: High-Resolution X-ray Structures of the Adduct Formed between Hen Egg-White Lysozyme and NAMI-A at Various Time Points.
Inorg.Chem., 60, 2021
|
|
7REU
 
 | | Crystal structure of Aro4p, 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase from Candida auris, L-Tyr complex | | Descriptor: | 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Tan, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-13 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of Aro4p, 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase from Candida auris, L-Tyr complex
To Be Published
|
|
7BDM
 
 | | The adduct of NAMI-A with Hen Egg White Lysozyme at 98 hours. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme, ... | | Authors: | Chiniadis, L, Giastas, P, Bratsos, I, Papakyriakou, A. | | Deposit date: | 2020-12-22 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Insights into the Protein Ruthenation Mechanism by Antimetastatic Metallodrugs: High-Resolution X-ray Structures of the Adduct Formed between Hen Egg-White Lysozyme and NAMI-A at Various Time Points.
Inorg.Chem., 60, 2021
|
|
5N3P
 
 | |
7BD0
 
 | | The adduct of NAMI-A with Hen Egg White Lysozyme at 26 hours. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Chiniadis, L, Giastas, P, Bratsos, I, Papakyriakou, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Insights into the Protein Ruthenation Mechanism by Antimetastatic Metallodrugs: High-Resolution X-ray Structures of the Adduct Formed between Hen Egg-White Lysozyme and NAMI-A at Various Time Points.
Inorg.Chem., 60, 2021
|
|
