5MI9
 
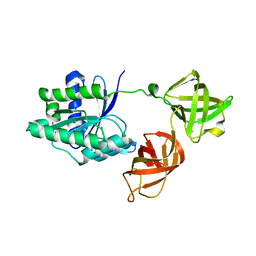 | | Structure of the phosphomimetic mutant of the elongation factor EF-Tu T62E | | Descriptor: | Elongation factor Tu 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Talavera, A, Hendrix, J, Versees, W, De Gieter, S, Castro-Roa, D, Jurenas, D, Van Nerom, K, Vandenberk, N, Barth, A, De Greve, H, Hofkens, J, Zenkin, N, Loris, R, Garcia-Pino, A. | | Deposit date: | 2016-11-27 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Phosphorylation decelerates conformational dynamics in bacterial translation elongation factors.
Sci Adv, 4, 2018
|
|
5MIX
 
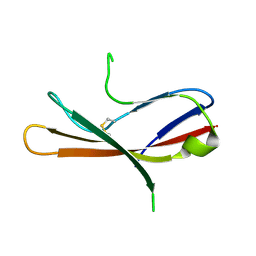 | |
5MJ5
 
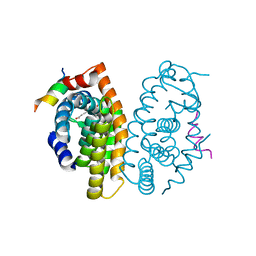 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetichonokiol derivative 3 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[4-oxidanyl-3-[3-(phenylmethyl)phenyl]phenyl]prop-2-enoic acid, LYS-HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN-ASP-SER, Retinoic acid receptor RXR-alpha | | Authors: | Andrei, S.A, Brunsveld, L, Scheepstra, M, Ottmann, C. | | Deposit date: | 2016-11-30 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
6PNP
 
 | | Crystal structure of the splice insert-free neurexin-1 LNS2 domain in complex with neurexophilin-1 | | Descriptor: | Neurexin-1, Neurexophilin-1 | | Authors: | Wilson, S.C, White, K.I, Zhou, Q, Brunger, A.T. | | Deposit date: | 2019-07-02 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of neurexophilin-neurexin complexes reveal a regulatory mechanism of alternative splicing.
Embo J., 38, 2019
|
|
5MFH
 
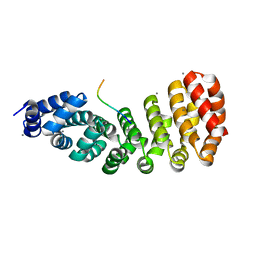 | | Designed armadillo repeat protein YIIIM5AII in complex with peptide (RR)5 | | Descriptor: | (RR)5, CALCIUM ION, YIIIM5AII | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
6PPM
 
 | | Ancestral Caspase 6 | | Descriptor: | Ancestral Caspase-6 Large Subunit, Ancestral Caspase-6 small subunit, VAL-GLU-ILE-ASP Inhibitor | | Authors: | Clark, A.C. | | Deposit date: | 2019-07-08 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Resurrection of ancestral effector caspases identifies novel networks for evolution of substrate specificity.
Biochem.J., 476, 2019
|
|
8CLS
 
 | |
6YD9
 
 | | Ecoli GyrB24 with inhibitor 16a | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, N-[6-(3-azanylpropanoylamino)-1,3-benzothiazol-2-yl]-3,4-bis(chloranyl)-5-methyl-1H-pyrrole-2-carboxamide | | Authors: | Barancokova, M, Skok, Z, Benek, O, Cruz, C.D, Tammela, P, Tomasic, T, Zidar, N, Masic, L.P, Zega, A, Stevenson, C.E.M, Mundy, J, Lawson, D.M, Maxwell, A.M, Kikelj, D, Ilas, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Exploring the Chemical Space of Benzothiazole-Based DNA Gyrase B Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
8CH5
 
 | |
8CA1
 
 | | Cryo-EM structure of the ACADVL dimer from Mus musculus. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Very long-chain specific acyl-CoA dehydrogenase, mitochondrial | | Authors: | Yin, Z, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2023-01-24 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insights into respiratory complex I deficiency and assembly from the mitochondrial disease-related ndufs4 -/- mouse.
Embo J., 43, 2024
|
|
8C40
 
 | | 14-3-3sigma bound to PKA-responsive ERa phosphopeptide | | Descriptor: | 14-3-3 protein sigma, Estrogen Receptor alpha phosphopeptide, MAGNESIUM ION | | Authors: | Somsen, B.A, Ottmann, C. | | Deposit date: | 2022-12-30 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis and dual ligand regulation of tetrameric estrogen receptor alpha /14-3-3 zeta protein complex.
J.Biol.Chem., 299, 2023
|
|
6Y1G
 
 | | Photoconverted HcRed in its optoacoustic state | | Descriptor: | 1,2-ETHANEDIOL, GFP-like non-fluorescent chromoprotein | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-02-12 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Challenging a Preconception: Optoacoustic Spectrum Differs from the Optical Absorption Spectrum of Proteins and Dyes for Molecular Imaging.
Anal.Chem., 92, 2020
|
|
1BB4
 
 | |
6Y2H
 
 | |
6UZS
 
 | |
6V52
 
 | | IDO1 IN COMPLEX WITH COMPOUND 1 | | Descriptor: | 3-chloro-N-{4-[1-(propylcarbamoyl)cyclobutyl]phenyl}benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Koenig, K.V, Augustin, M.A. | | Deposit date: | 2019-12-03 | | Release date: | 2020-04-08 | | Last modified: | 2020-04-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Strategic Incorporation of Polarity in Heme-Displacing Inhibitors of Indoleamine-2,3-dioxygenase-1 (IDO1).
Acs Med.Chem.Lett., 11, 2020
|
|
6YPS
 
 | | Crystal structure of the cAMP-dependent protein kinase A in complex with 4-hydroxybenzamidine | | Descriptor: | 4-oxidanylbenzenecarboximidamide, DIMETHYL SULFOXIDE, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Oebbeke, M, Siefker, C, Heine, A, Klebe, G. | | Deposit date: | 2020-04-16 | | Release date: | 2020-12-09 | | Last modified: | 2020-12-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Fragment Binding to Kinase Hinge: If Charge Distribution and Local pK a Shifts Mislead Popular Bioisosterism Concepts.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6V61
 
 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in the Complex with the Inhibitor Captopril | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in the Complex with the Inhibitor Captopril.
To Be Published
|
|
8CJL
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor TPT-004 | | Descriptor: | 3-ethyl-8-[(2-methyl-5~{H}-imidazo[2,1-b][1,3]thiazol-6-yl)methyl]-7-(oxetan-3-ylmethyl)purine-2,6-dione, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure-Based Design of Xanthine-Imidazopyridines and -Imidazothiazoles as Highly Potent and In Vivo Efficacious Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
6Y58
 
 | | Binary complex of 14-3-3 sigma (C38N) with the Estrogen Related Receptor gamma (LBD) phosphopeptide | | Descriptor: | 14-3-3 protein sigma, Estrogen Related Receptor gamma phosphopeptide, MAGNESIUM ION | | Authors: | Somsen, B.A, Sijbesma, E, Leijten-van de Gevel, I.A, Ottmann, C. | | Deposit date: | 2020-02-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Fluorescence Anisotropy-Based Tethering for Discovery of Protein-Protein Interaction Stabilizers.
Acs Chem.Biol., 15, 2020
|
|
6VA1
 
 | | Solution Structure of the Tau pre-mRNA Exon 10 Splicing Regulatory Element | | Descriptor: | RNA (5'-R(*CP*AP*CP*AP*CP*GP*UP*CP*GP*G)-3'), RNA (5'-R(*CP*CP*GP*GP*CP*AP*GP*UP*GP*UP*G)-3') | | Authors: | Chen, J.L, Fountain, M.A, Disney, M.D. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Design, Optimization, and Study of Small Molecules That Target Tau Pre-mRNA and Affect Splicing.
J.Am.Chem.Soc., 142, 2020
|
|
6V1U
 
 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to TP-472 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-acetylpyrrolo[1,2-a]pyrimidin-8-yl)-N-cyclopropyl-4-methylbenzamide, Bromodomain-containing protein 4 | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-21 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
5MI3
 
 | | Structure of phosphorylated translation elongation factor EF-Tu from E. coli | | Descriptor: | Elongation factor Tu 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Talavera, A, Hendrix, J, Versees, W, De Gieter, S, Castro-Roa, D, Jurenas, D, Van Nerom, K, Vandenberk, N, Barth, A, De Greve, H, Hofkens, J, Zenkin, N, Loris, R, Garcia-Pino, A. | | Deposit date: | 2016-11-27 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phosphorylation decelerates conformational dynamics in bacterial translation elongation factors.
Sci Adv, 4, 2018
|
|
6MVJ
 
 | | Cellobiose complex Cel45A from Neurospora crassa OR74A | | Descriptor: | Endoglucanase V, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kadowaki, M.A.S, Polikarpov, I. | | Deposit date: | 2018-10-25 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.809 Å) | | Cite: | Structural insights into the hydrolysis pattern and molecular dynamics simulations of GH45 subfamily a endoglucanase from Neurospora crassa OR74A.
Biochimie, 165, 2019
|
|
6V2B
 
 | |
