6MQI
 
 | |
8TDQ
 
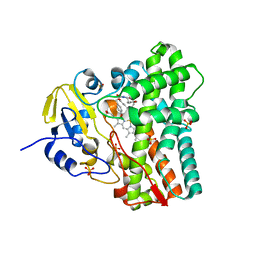 | | SFX-XFEL structure of CYP121 cocrystallized with substrate cYY | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Nguyen, R.C, Dasgupta, M, Bhowmick, A, Kern, J.F, Liu, A. | | Deposit date: | 2023-07-04 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | In Situ Structural Observation of a Substrate- and Peroxide-Bound High-Spin Ferric-Hydroperoxo Intermediate in the P450 Enzyme CYP121.
J.Am.Chem.Soc., 145, 2023
|
|
6XLR
 
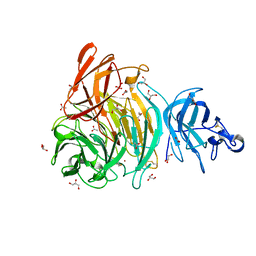 | |
8TSD
 
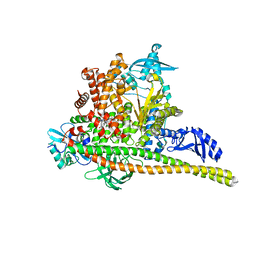 | | Human PI3K p85alpha/p110alpha bound to RLY-2608 | | Descriptor: | N-{(3R,6M)-3-(2-chloro-5-fluorophenyl)-6-[(4S)-5-cyano[1,2,4]triazolo[1,5-a]pyridin-6-yl]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}-3-fluoro-5-(trifluoromethyl)benzamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
6MQZ
 
 | |
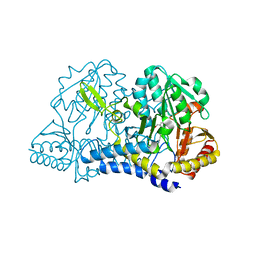
6MR6
 
 | |
8TS7
 
 | | Human PI3K p85alpha/p110alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
7AY8
 
 | | NMR solution structure of Tbo-IT2 | | Descriptor: | Tbo-IT2 | | Authors: | Mineev, K.S, Kornilov, F.D, Lushpa, V.A, Korolkova, Y.A, Maleeva, E.E, Andreev, Y.A, Kozlov, S.A, Arseniev, A.S. | | Deposit date: | 2020-11-11 | | Release date: | 2021-03-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | New Insectotoxin from Tibellus Oblongus Spider Venom Presents Novel Adaptation of ICK Fold.
Toxins, 13, 2021
|
|
4YHH
 
 | | Crystal structure of the 30S ribosomal subunit from Thermus thermophilus in complex with tigecycline | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Kaminishi, T, Ochoa-Lizarralde, B, Dhimole, N, Zhou, S, Lopez-Alonso, J.P, Connell, S.R, Fucini, P. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.417 Å) | | Cite: | Structural characterization of an alternative mode of tigecycline binding to the bacterial ribosome.
Antimicrob.Agents Chemother., 59, 2015
|
|
8TDP
 
 | | Time-resolved SFX-XFEL crystal structure of CYP121 bound with cYY reacted with peracetic acid for 200 milliseconds | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, HYDROGEN PEROXIDE, Mycocyclosin synthase, ... | | Authors: | Nguyen, R.C, Dasgupta, M, Bhowmick, A, Kern, J.F, Liu, A. | | Deposit date: | 2023-07-04 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In Situ Structural Observation of a Substrate- and Peroxide-Bound High-Spin Ferric-Hydroperoxo Intermediate in the P450 Enzyme CYP121.
J.Am.Chem.Soc., 145, 2023
|
|
7OJX
 
 | | E2 UBE2K covalently linked to donor Ub, acceptor di-Ub, and RING E3 primed for K48-linked Ub chain synthesis | | Descriptor: | 1,1'-ethane-1,2-diylbis(1H-pyrrole-2,5-dione), E3 ubiquitin-protein ligase RNF38, Polyubiquitin-B, ... | | Authors: | Majorek, K.A, Nakasone, M.A, Huang, D.T. | | Deposit date: | 2021-05-17 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of UBE2K-Ub/E3/polyUb reveals mechanisms of K48-linked Ub chain extension.
Nat.Chem.Biol., 18, 2022
|
|
7BT8
 
 | | Mevo lectin complex with mannotriose | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose, ... | | Authors: | Sivaji, N, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 2020-03-31 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and related studies on Mevo lectin from Methanococcus voltae A3: the first thorough characterization of an archeal lectin and its interactions.
Glycobiology, 31, 2021
|
|
6XU4
 
 | | Crystal structure of the genetically-encoded FGCaMP calcium indicator in its calcium-bound state | | Descriptor: | CALCIUM ION, FGCamp | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Barykina, N.V, Subach, O.M, Subach, F.V. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | FGCaMP7, an Improved Version of Fungi-Based Ratiometric Calcium Indicator for In Vivo Visualization of Neuronal Activity.
Int J Mol Sci, 21, 2020
|
|
8TS9
 
 | | Human PI3K p85alpha/p110alpha H1047R bound to compound 1 | | Descriptor: | 5-[3-fluoro-5-(trifluoromethyl)benzamido]-N-methyl-6-(2-methylanilino)pyridine-3-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8TSC
 
 | | Human PI3K p85alpha/p110alpha H1047R bound to compound 3 | | Descriptor: | (1S)-7-[3-fluoro-5-(trifluoromethyl)benzamido]-N-methyl-1-(2-methylphenyl)-3-oxo-2,3-dihydro-1H-isoindole-5-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8PFR
 
 | | Mouse RPL39L integrated into the yeast 60S ribosomal subunit | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | Deposit date: | 2023-06-16 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Mouse RPL39L integrated into the yeast 60S ribosomal subunit
To Be Published
|
|
7NUY
 
 | |
7QTO
 
 | |
8F4P
 
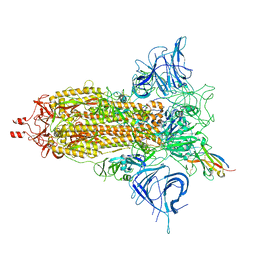 | |
6XKP
 
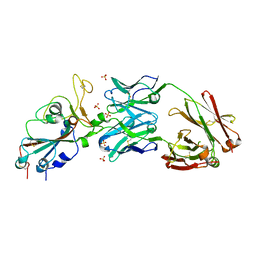 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CV07-270 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV07-270 Heavy Chain, CV07-270 Light Chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.
Cell, 183, 2020
|
|
8F7H
 
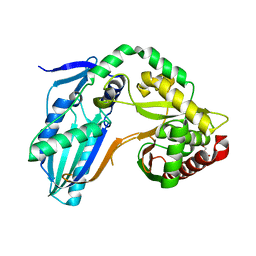 | | The condensation domain of surfactin A synthetase C variant 18b in space group P212121 | | Descriptor: | GLYCEROL, Surfactin synthetase | | Authors: | Frota, N.F, Pistofidis, A, Folger, I.B, Hilvert, D, Schmeing, M. | | Deposit date: | 2022-11-18 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | High-throughput reprogramming of an NRPS condensation domain.
Nat.Chem.Biol., 20, 2024
|
|
7QTU
 
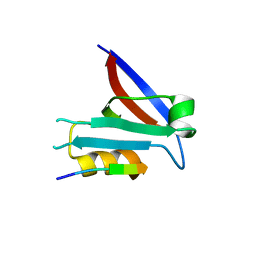 | |
8F7I
 
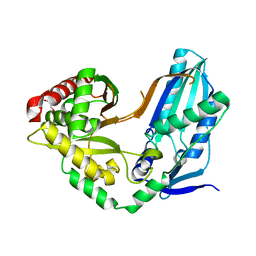 | | The condensation domain of surfactin A synthetase C variant 18b in space group P43212 | | Descriptor: | GLYCEROL, Surfactin synthetase | | Authors: | Frota, N.F, Pistofidis, A, Folger, I.B, Hilvert, D, Schmeing, M. | | Deposit date: | 2022-11-18 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | High-throughput reprogramming of an NRPS condensation domain.
Nat.Chem.Biol., 20, 2024
|
|
7OFY
 
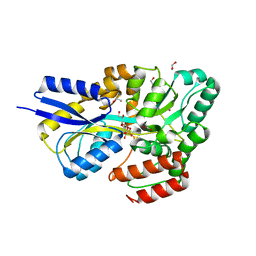 | | Crystal structure of SQ binding protein from Agrobacterium tumefaciens in complex with sulfoquinovosyl glycerol (SQGro) | | Descriptor: | 1,2-ETHANEDIOL, Sulfoquinovosyl binding protein, [(2S,3S,4S,5R,6S)-6-[(2R)-2,3-bis(oxidanyl)propoxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Jarva, M.A, Sharma, M, Goddard-Borger, E.D, Davies, G.J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oxidative desulfurization pathway for complete catabolism of sulfoquinovose by bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8F7G
 
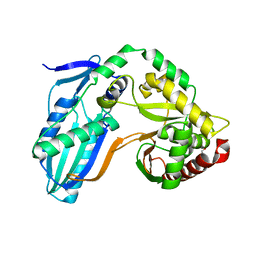 | | The condensation domain of surfactin A synthetase C in space group P212121 | | Descriptor: | GLYCEROL, Surfactin synthetase | | Authors: | Frota, N.F, Pistofidis, A, Folger, I.B, Hilvert, D, Schmeing, T.M. | | Deposit date: | 2022-11-18 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-throughput reprogramming of an NRPS condensation domain.
Nat.Chem.Biol., 20, 2024
|
|
