5ML7
 
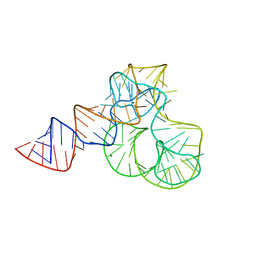 | | CRYSTAL STRUCTURE OF L1-STALK FRAGMENT OF 23S rRNA FROM HALOARCULA MARISMORTUI | | Descriptor: | 23S ribosomal RNA, CALCIUM ION, MAGNESIUM ION | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Nevskaya, N.A, Nikonov, S.V. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the 23S rRNA Fragment Specific to r-Protein L1 and Designed Model of the Ribosomal L1 Stalk from Haloarcula marismortui
Crystals, 2017
|
|
6OZZ
 
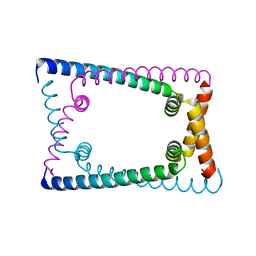 | | N terminally deleted GapR crystal structure from C. crescentus | | Descriptor: | UPF0335 protein CC_3319 | | Authors: | Tarry, M, Harmel, C, Taylor, J.A, Marczynski, G.T, Schmeing, T.M. | | Deposit date: | 2019-05-16 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.304 Å) | | Cite: | Structures of GapR reveal a central channel which could accommodate B-DNA.
Sci Rep, 9, 2019
|
|
6P0A
 
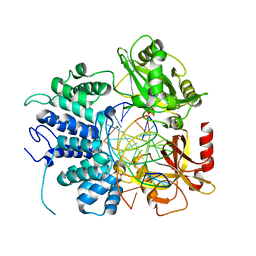 | | Human DNA Ligase 1 Bound to an Adenylated, dideoxy Terminated DNA nick with 2 mM Mg2+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*C)-3'), DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Schellenberg, M.J, Williams, R.S, Tumbale, P.S, Riccio, A.A. | | Deposit date: | 2019-05-16 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Two-tiered enforcement of high-fidelity DNA ligation.
Nat Commun, 10, 2019
|
|
6PBK
 
 | |
6P3H
 
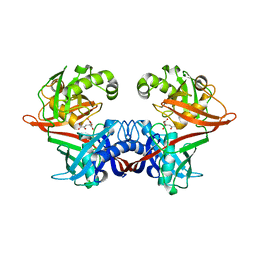 | | Crystal structure of LigU(K66M) bound to substrate | | Descriptor: | (1E)-4-oxobut-1-ene-1,2,4-tricarboxylic acid, (4E)-oxalomesaconate Delta-isomerase, CHLORIDE ION | | Authors: | Cory, S.A, Hogancamp, T.N, Raushel, F.M, Barondeau, D.P. | | Deposit date: | 2019-05-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure and Chemical Reaction Mechanism of LigU, an Enzyme That Catalyzes an Allylic Isomerization in the Bacterial Degradation of Lignin.
Biochemistry, 58, 2019
|
|
6P45
 
 | |
6MPK
 
 | |
6PF1
 
 | |
4UJ6
 
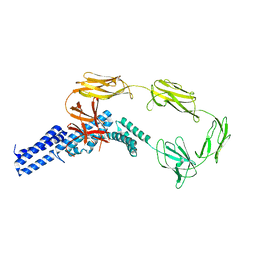 | | Structure of surface layer protein SbsC, domains 1-6 | | Descriptor: | SURFACE LAYER PROTEIN | | Authors: | Dordic, A, Pavkov-Keller, T, Eder, M, Egelseer, E.M, Davis, K, Mills, D, Sleytr, U.B, Kuehlbrandt, W, Vonck, J, Keller, W. | | Deposit date: | 2015-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of Surface Layer Protein Sbsc, Domains 1-6
To be Published
|
|
6MQY
 
 | |
7NLV
 
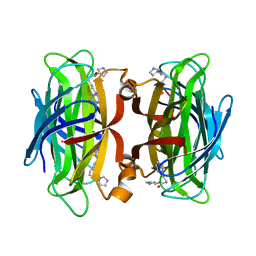 | | WILDTYPE CORE-STREPTAVIDIN WITH a conjugated BIOTINYLATED PYRROLIDINE II | | Descriptor: | 5-((3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)-N-((S)-pyrrolidin-3-yl)pentanamide, Streptavidin | | Authors: | Nodling, A.R, Santi, N, Tsai, Y.H, Rizkallah, P, Luk, L.Y.P, Jin, Y. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The role of streptavidin and its variants in catalysis by biotinylated secondary amines.
Org.Biomol.Chem., 19, 2021
|
|
6MRC
 
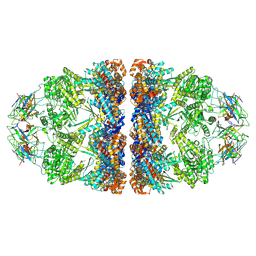 | | ADP-bound human mitochondrial Hsp60-Hsp10 football complex | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Gomez-Llorente, Y, Jebara, F, Patra, M, Malik, R, Nissemblat, S, Azem, A, Hirsch, J.A, Ubarretxena-Belandia, I. | | Deposit date: | 2018-10-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for active single and double ring complexes in human mitochondrial Hsp60-Hsp10 chaperonin.
Nat Commun, 11, 2020
|
|
8C29
 
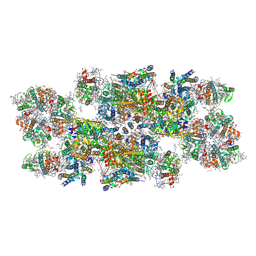 | | Cryo-EM structure of photosystem II C2S2 supercomplex from Norway spruce (Picea abies) at 2.8 Angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Kopecny, D, Semchonok, D.A, Kouril, R. | | Deposit date: | 2022-12-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.785 Å) | | Cite: | Cryo-EM structure of a plant photosystem II supercomplex with light-harvesting protein Lhcb8 and alpha-tocopherol.
Nat.Plants, 9, 2023
|
|
7AWF
 
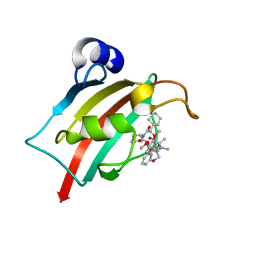 | | The Fk1 domain of FKBP51 in complex with (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15,16-trimethyl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^5,^10]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone | | Descriptor: | (2~{R},5~{S},12~{R})-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15,16-trimethyl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^{5,10}]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Voll, M.A, Meyners, C, Heymann, T, Merz, S, Purder, P, Bracher, A, Hausch, F. | | Deposit date: | 2020-11-07 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Macrocyclic FKBP51 Ligands Define a Transient Binding Mode with Enhanced Selectivity.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7AOU
 
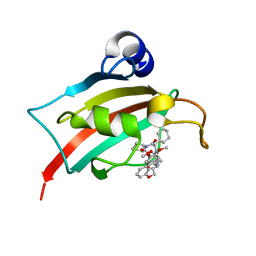 | | The Fk1 domain of FKBP51 in complex with (2'R,5'S,12'R)-12'-cyclohexyl-2'-[2-(3,4-dimethoxyphenyl)ethyl]-3',19'-dioxa-10',13',16'-triazaspiro[cyclopropane-1,15'- tricyclo[18.3.1.0-5,10]tetracosane]-1'(24'),20',22'-triene-4',11',14',17'-tetrone | | Descriptor: | (2'R,5'S,12'R)-12'-cyclohexyl-2'-[2-(3,4-dimethoxyphenyl)ethyl]-3',19'-dioxa-10',13',16'-triazaspiro[cyclopropane-1,15'- tricyclo[18.3.1.0-5,10]tetracosane]-1'(24'),20',22'-triene-4',11',14',17'-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Voll, M.A, Meyners, C, Heymann, T, Merz, S, Purder, P, Bracher, A, Hausch, F. | | Deposit date: | 2020-10-15 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Macrocyclic FKBP51 Ligands Define a Transient Binding Mode with Enhanced Selectivity.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6UTK
 
 | |
7ASE
 
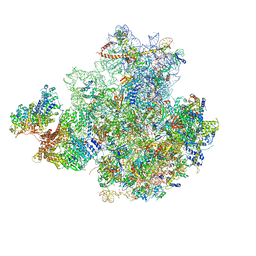 | | 43S preinitiation complex from Trypanosoma cruzi with the kDDX60 helicase | | Descriptor: | 18S, 40S ribosomal protein S10, putative, ... | | Authors: | Bochler, A, Brito Querido, J, Prilepskaja, T, Soufari, H, Del Cistia, M.L, Kuhn, L, Rimoldi Ribeiro, A, Valasek, L.S, Hashem, Y. | | Deposit date: | 2020-10-27 | | Release date: | 2021-04-14 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural Differences in Translation Initiation between Pathogenic Trypanosomatids and Their Mammalian Hosts.
Cell Rep, 33, 2020
|
|
7POA
 
 | | An Irreversible, Promiscuous and Highly Thermostable Claisen-Condensation Biocatalyst Drives the Synthesis of Substituted Pyrroles | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-amino-7-oxononanoate synthase/2-amino-3-ketobutyrate coenzyme A ligase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Basle, A, Ashley, B, Campopiano, D, Marles-Wright, J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Versatile Chemo-Biocatalytic Cascade Driven by a Thermophilic and Irreversible C-C Bond-Forming alpha-Oxoamine Synthase.
Acs Sustain Chem Eng, 11, 2023
|
|
8CMC
 
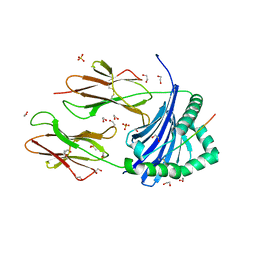 | | Human Leukocyte Antigen class II allotype DR1 presenting SARS-CoV-2 Spike peptide S511-530 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HLA class II histocompatibility antigen, ... | | Authors: | MacLachlan, B.J, Mason, G.H, Sourfield, D.O, Godkin, A.J, Rizkallah, P.J. | | Deposit date: | 2023-02-19 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural definition of HLA class II-presented SARS-CoV-2 epitopes reveals a mechanism to escape pre-existing CD4 + T cell immunity.
Cell Rep, 42, 2023
|
|
7POB
 
 | | An Irreversible, Promiscuous and Highly Thermostable Claisen-Condensation Biocatalyst Drives the Synthesis of Substituted Pyrroles | | Descriptor: | 8-amino-7-oxononanoate synthase/2-amino-3-ketobutyrate coenzyme A ligase, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION | | Authors: | Basle, A, Ashley, B, Campopiano, D, Marles-Wright, J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Versatile Chemo-Biocatalytic Cascade Driven by a Thermophilic and Irreversible C-C Bond-Forming alpha-Oxoamine Synthase.
Acs Sustain Chem Eng, 11, 2023
|
|
8CMG
 
 | | Human Leukocyte Antigen class II allotype DR1 presenting SARS-CoV-2 nsp14 peptide (orf1ab)6420-6434 | | Descriptor: | 1,2-ETHANEDIOL, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | MacLachlan, B.J, Mason, G.H, Sourfield, D.O, Godkin, A.J, Rizkallah, P.J. | | Deposit date: | 2023-02-19 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural definition of HLA class II-presented SARS-CoV-2 epitopes reveals a mechanism to escape pre-existing CD4 + T cell immunity.
Cell Rep, 42, 2023
|
|
7POC
 
 | | An Irreversible, Promiscuous and Highly Thermostable Claisen-Condensation Biocatalyst Drives the Synthesis of Substituted Pyrroles | | Descriptor: | 8-amino-7-oxononanoate synthase/2-amino-3-ketobutyrate coenzyme A ligase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Basle, A, Ashley, B, Campopiano, D, Marles-Wright, J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Versatile Chemo-Biocatalytic Cascade Driven by a Thermophilic and Irreversible C-C Bond-Forming alpha-Oxoamine Synthase.
Acs Sustain Chem Eng, 11, 2023
|
|
6Y0N
 
 | | Arginine hydroxylase VioC in complex with Arg, 2OG and Fe under anaerobic environment using FT-SSX methods | | Descriptor: | 2-OXOGLUTARIC ACID, ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, ... | | Authors: | Rabe, P, Beale, J.H, Lang, P.A, Dirr, S.A, Leissing, T.M, Butryn, A, Aller, P, Kamps, J.J.A.G, Axford, D, McDonough, M.A, Orville, A.M, Owen, R, Schofield, C.J. | | Deposit date: | 2020-02-10 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Anaerobic fixed-target serial crystallography.
Iucrj, 7, 2020
|
|
6P3T
 
 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with inhibitor SGT449 | | Descriptor: | AMMONIUM ION, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2019-05-24 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the Robustness of Inhibitors of Tuberculosis Aminoglycoside Resistance Enzyme Eis by Mutagenesis.
Acs Infect Dis., 5, 2019
|
|
8F8X
 
 | | Crystal structure of Nb.X0 bound to the afucosylated human IgG1 fragment crystal form II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb.X0, Uncharacterized protein DKFZp686C11235 | | Authors: | Goldgur, Y, Ravetch, J, Gupta, A, Kao, K, Oren, D. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of glycoform specificity and in vivo protection by an anti-afucosylated IgG nanobody.
Nat Commun, 14, 2023
|
|
