5ZKH
 
 | |
7P5M
 
 | | Cryo-EM structure of human TTYH2 in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 2 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
7P5C
 
 | | Cryo-EM structure of human TTYH3 in Ca2+ and GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 3 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
5KT0
 
 | | Dihydrodipicolinate reductase from the industrial and evolutionarily important cyanobacteria Anabaena variabilis. | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate reductase, MAGNESIUM ION | | Authors: | Christensen, J.B, Soares da Costa, T.P, Faou, P, Pearce, F.G, Panjikar, S, Perugini, M.A. | | Deposit date: | 2016-07-10 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Dihydrodipicolinate Reductase from the industrial and evolutionarily important cyanobacteria Anabaena variabilis.
To Be Published
|
|
7P54
 
 | | Cryo-EM structure of human TTYH2 in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 2 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
7P5J
 
 | | Cryo-EM structure of human TTYH1 in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 1 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
8BWH
 
 | |
8BWE
 
 | |
8BTQ
 
 | |
8C1Y
 
 | | Small molecule stabilizer for 14-3-3/ChREBP (Cmd 30) | | Descriptor: | 14-3-3 protein sigma, Carbohydrate-responsive element-binding protein, [2-[2-[[2,2-bis(fluoranyl)-2-phenyl-ethyl]amino]-2-oxidanylidene-ethoxy]phenyl]phosphonic acid | | Authors: | Pennings, M.A.M, Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-21 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular glues of the regulatory ChREBP/14-3-3 complex protect beta cells from glucolipotoxicity.
Biorxiv, 2024
|
|
8SWK
 
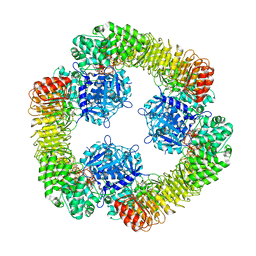 | | Cryo-EM structure of NLRP3 closed hexamer | | Descriptor: | 1-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-3-(1,2,3,5-tetrahydro-s-indacen-4-yl)urea, ADENOSINE-5'-TRIPHOSPHATE, NACHT, ... | | Authors: | Yu, X, Matico, R.E, Miller, R, Schoubroeck, B.V, Grauwen, K, Suarez, J, Pietrak, B, Haloi, N, Yin, Y, Tresadern, G.J, Perez-Benito, L, Lindahl, E, Bottelbergs, A, Oehlrich, D, Opdenbosch, N.V, Sharma, S. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Cryo-EM structures of NLRP3 reveal its self-activation mechanism
Nat Commun, 2024
|
|
6O1Y
 
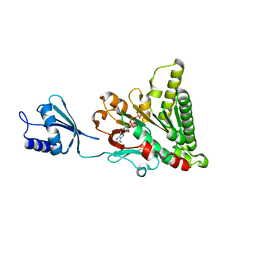 | | Structure of pCW3 conjugation coupling protein TcpA monomeric form with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA translocase coupling protein, beta-D-glucopyranose | | Authors: | Traore, D.A.K, Ahktar, N, Torres, V.T, Adams, V, Coulibaly, F, Panjikar, S, Caradoc-Davies, T.T, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2019-02-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of pCW3 conjugation coupling protein TcpA
To be published
|
|
7QO9
 
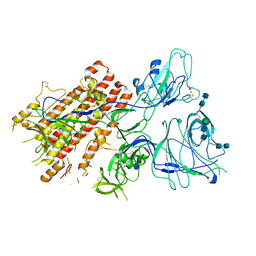 | | SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD and NTD (Local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,SARS-CoV-2 S Omicron Spike B.1.1.529, ... | | Authors: | Ni, D, Lau, K, Turelli, P, Beckert, B, Nazarov, S, Pojer, F, Myasnikov, A, Stahlberg, H, Trono, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structural analysis of the Spike of the Omicron SARS-COV-2 variant by cryo-EM and implications for immune evasion
Biorxiv, 2021
|
|
7QGJ
 
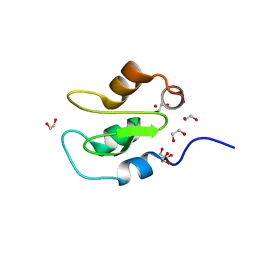 | | Apo structure of BIR2 Domain of BIRC2 | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Kraemer, A, Farges, F, Schwalm, M.P, Saxena, K, Preuss, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-12-08 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Apo structure BIR2 Domain of BIRC2
To Be Published
|
|
1BTT
 
 | | THE SOLUTION STRUCTURES OF THE FIRST AND SECOND TRANSMEMBRANE-SPANNING SEGMENTS OF BAND 3 | | Descriptor: | BAND 3 ANION TRANSPORT PROTEIN | | Authors: | Gargaro, A.R, Bloomberg, G.B, Dempsey, C.E, Murray, M, Tanner, M.J.A. | | Deposit date: | 1994-08-03 | | Release date: | 1994-12-20 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the first and second transmembrane-spanning segments of band 3.
Eur.J.Biochem., 221, 1994
|
|
1BTR
 
 | | THE SOLUTION STRUCTURES OF THE FIRST AND SECOND TRANSMEMBRANE-SPANNING SEGMENTS OF BAND 3 | | Descriptor: | BAND 3 ANION TRANSPORT PROTEIN | | Authors: | Gargaro, A.R, Bloomberg, G.B, Dempsey, C.E, Murray, M, Tanner, M.J.A. | | Deposit date: | 1993-05-25 | | Release date: | 1994-12-20 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the first and second transmembrane-spanning segments of band 3.
Eur.J.Biochem., 221, 1994
|
|
5L8P
 
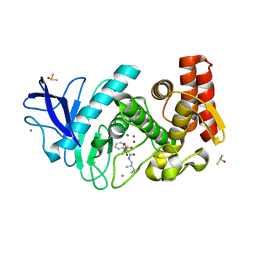 | | Thermolysin in complex with JC114 (PEG400 cryo protectant) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, N~2~-[(R)-({[(benzyloxy)carbonyl]amino}methyl)(hydroxy)phosphoryl]-N-[(2S)-2,3,3-trimethylbutyl]-L-leucinamide, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-06-08 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Rational Design of Thermodynamic and Kinetic Binding Profiles by Optimizing Surface Water Networks Coating Protein-Bound Ligands.
J. Med. Chem., 59, 2016
|
|
6NR5
 
 | | Human LSD1 in complex with Phenelzine sulfate | | Descriptor: | Lysine-specific histone demethylase 1A, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3R,4R)-5-[(4aR)-7,8-dimethyl-2,4-dioxo-5-(2-phenylethyl)-3,4,4a,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Tan, A.H.Y, Tu, W, McCuaig, R, Donovan, T, Tsimbalyuk, S, Forwood, J.K, Rao, S. | | Deposit date: | 2019-01-22 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Lysine-Specific Histone Demethylase 1A Regulates Macrophage Polarization and Checkpoint Molecules in the Tumor Microenvironment of Triple-Negative Breast Cancer.
Front Immunol, 10, 2019
|
|
6O1W
 
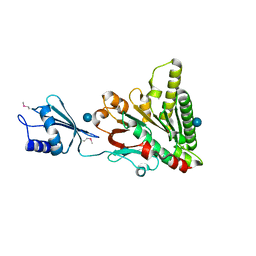 | | Structure of pCW3 conjugation coupling protein TcpA monomer orthorhombic crystal form | | Descriptor: | DNA translocase coupling protein, beta-D-glucopyranose | | Authors: | Traore, D.A.K, Ahktar, N, Torres, V.T, Adams, V, Coulibaly, F, Panjikar, S, Caradoc-Davies, T.T, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2019-02-22 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of pCW3 conjugation coupling protein TcpA
To be published
|
|
7ZWD
 
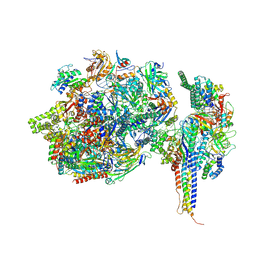 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U5 snRNA promoter (CC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZX7
 
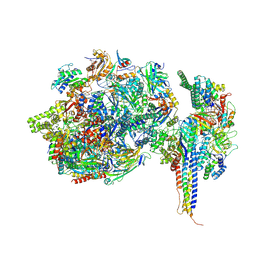 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (CC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5L8F
 
 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E32A, E62A, H65A. | | Descriptor: | MAGNESIUM ION, Rru_A0973, TRIETHYLENE GLYCOL | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E32A, E62A, H65A.
To be published
|
|
7ZXE
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (OC) | | Descriptor: | Non-template strand, TATA-box-binding protein, Template strand, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6NJK
 
 | |
7OJT
 
 | | Crystal structure of unliganded PatA, a membrane associated acyltransferase from Mycobacterium smegmatis | | Descriptor: | GLYCEROL, Phosphatidylinositol mannoside acyltransferase | | Authors: | Anso, I, Wang, L, Marina, A, Paez-Perez, E.D, Perrone, S, Lowary, T.L, Trastoy, B, Guerin, M.E. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.67 Å) | | Cite: | Molecular ruler mechanism and interfacial catalysis of the integral membrane acyltransferase PatA.
Sci Adv, 7, 2021
|
|
