3KKY
 
 | | Structure of Manganese Superoxide Dismutase from Deinococcus Radiodurans in the orthorhombic space group P212121: A case study of mistaken identity | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn] | | Authors: | Govindasamy, L, Mikulski, R, McKenna, M.A, Silverman, D.N, McKenna, R. | | Deposit date: | 2009-11-06 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Manganese Superoxide dismutase from Deinococcus Radiodurans in the orthorhombic space group P212121: A case study of mistaken identity
To be Published
|
|
1RBQ
 
 | | Human GAR Tfase complex structure with 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid | | Descriptor: | N-{4-[(1R)-4-[(2R,4R,5S)-2,4-DIAMINO-6-OXOHEXAHYDROPYRIMIDIN-5-YL]-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXYETHYL)BUTYL]BENZOYL}-D-GLUTAMIC ACID, PHOSPHATE ION, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Zhang, Y, Desharnais, J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-11-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Human GAR Tfase complex structure with polyglutamated
10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid
To be Published
|
|
5VVL
 
 | | Cas1-Cas2 bound to full-site mimic with Ni | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (11-MER), ... | | Authors: | Wright, A.V, Knott, G.J, Doxzen, K.D, Doudna, J.A. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structures of the CRISPR genome integration complex.
Science, 357, 2017
|
|
5YX1
 
 | | Crystal structure of hematopoietic prostaglandin D synthase in complex with U004 | | Descriptor: | GLUTATHIONE, GLYCEROL, Hematopoietic prostaglandin D synthase, ... | | Authors: | Kamo, M, Furubayashi, N, Inaka, K, Aritake, K, Urade, Y, Takaya, D, Tanaka, A. | | Deposit date: | 2017-12-01 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal structure of hematopoietic prostaglandin D synthase in complex with U004
To Be Published
|
|
2X2O
 
 | | The flavoprotein NrdI from Bacillus cereus with the initially oxidized FMN cofactor in an intermediate radiation reduced state | | Descriptor: | CACODYLATE ION, FLAVIN MONONUCLEOTIDE, NRDI PROTEIN, ... | | Authors: | Rohr, A.K, Hersleth, H.P, Andersson, K.K. | | Deposit date: | 2010-01-15 | | Release date: | 2010-03-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Tracking Flavin Conformations in Protein Crystal Structures with Raman Spectroscopy and Qm/Mm Calculations
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
7KD3
 
 | |
3R9K
 
 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, glu47asp mutant complexed with sulfate, a closed cap conformation | | Descriptor: | Putative beta-phosphoglucomutase, SULFATE ION | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-03-25 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
5VQF
 
 | | Crystal Structure of pro-TGF-beta 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Transforming growth factor beta-1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zhao, B, Xu, S, Dong, X, Lu, C, Springer, T.A. | | Deposit date: | 2017-05-08 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Prodomain-growth factor swapping in the structure of pro-TGF-beta 1.
J. Biol. Chem., 293, 2018
|
|
2XT1
 
 | | Crystal structure of the HIV-1 capsid protein C-terminal domain (146- 231) in complex with a camelid VHH. | | Descriptor: | CAMELID VHH 9, GAG POLYPROTEIN, GLYCEROL | | Authors: | Igonet, S, Vaney, M.C, Bartonova, V, Helma, J, Rothbauer, U, Leonhardt, H, Stura, E, Krausslich, H.-G, Rey, F.A. | | Deposit date: | 2010-10-03 | | Release date: | 2011-10-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Targeting HIV-1 Virion Formation with Nanobodies -Implications for the Design of Assembly Inhibitors
To be Published
|
|
1LV1
 
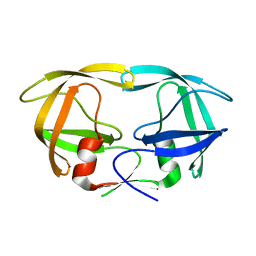 | | Crystal Structure Analysis of the non-active site mutant of tethered HIV-1 protease to 2.1A resolution | | Descriptor: | HIV-1 protease | | Authors: | Kumar, M, Kannan, K.K, Hosur, M.V, Bhavesh, N.S, Chatterjee, A, Mittal, R, Hosur, R.V. | | Deposit date: | 2002-05-24 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effects of remote mutation on the autolysis of HIV-1 PR: X-ray and NMR investigations.
Biochem.Biophys.Res.Commun., 294, 2002
|
|
5H83
 
 | | HETEROYOHIMBINE SYNTHASE HYS FROM CATHARANTHUS ROSEUS - APO FORM | | Descriptor: | ZINC ION, heteroyohimbine synthase HYS | | Authors: | Stavrinides, A, Tatsis, E.C, Caputi, L, Foureau, E, Stevenson, C.E.M, Lawson, D.M, Courdavault, V, O'Connor, S.E. | | Deposit date: | 2015-12-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural investigation of heteroyohimbine alkaloid synthesis reveals active site elements that control stereoselectivity.
Nat Commun, 7, 2016
|
|
4AP8
 
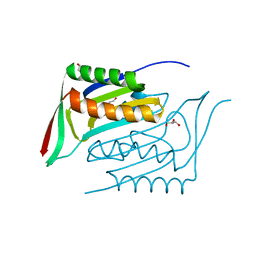 | | Crystal structure of human Molybdopterin synthase catalytic subunit (MOCS2B) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MOLYBDOPTERIN SYNTHASE CATALYTIC SUBUNIT | | Authors: | Vollmar, M, Kiyani, W, Krojer, T, Goubin, S, Allerston, C, Froese, D.S, von Delft, F, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Yue, W.W. | | Deposit date: | 2012-03-30 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal Structure of Human Molybdopterin Synthase Catalytic Subunit (Mocs2B)
To be Published
|
|
5VS1
 
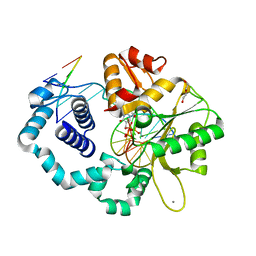 | |
3AM7
 
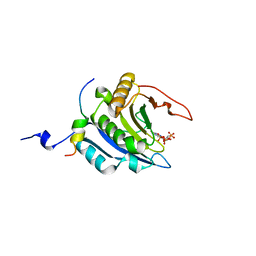 | | Crystal structure of the ternary complex of eIF4E-M7GTP-4EBP2 peptide | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4E-binding protein 2 | | Authors: | Tomoo, K, Fukuyo, A, In, Y, Ishida, T. | | Deposit date: | 2010-08-17 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural scaffold for eIF4E binding selectivity of 4E-BP isoforms: crystal structure of eIF4E binding region of 4E-BP2 and its comparison with that of 4E-BP1.
J.Pept.Sci., 17, 2011
|
|
5HBQ
 
 | | C63D mutant of the rhodanese domain of YgaP | | Descriptor: | CHLORIDE ION, Inner membrane protein YgaP, SODIUM ION | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | Deposit date: | 2016-01-02 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
3KTA
 
 | |
3RMO
 
 | | Human Thrombin in complex with MI004 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ligand binding stepwise disrupts water network in thrombin: enthalpic and entropic changes reveal classical hydrophobic effect
J.Med.Chem., 55, 2012
|
|
3KT9
 
 | | Aprataxin FHA Domain | | Descriptor: | Aprataxin | | Authors: | Cherry, A.L, Smerdon, S.J. | | Deposit date: | 2009-11-24 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | CK2 phosphorylation-dependent interaction between aprataxin and MDC1 in the DNA damage response.
Nucleic Acids Res., 38, 2010
|
|
7Z79
 
 | | Crystal structure of aminotransferase-like protein from Variovorax paradoxus | | Descriptor: | Aminotransferase, class 4, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Boyko, K.M, Matyuta, I.O, Nikolaeva, A.Y, Khrenova, M.G, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-03-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Puzzling Protein from Variovorax paradoxus Has a PLP Fold Type IV Transaminase Structure and Binds PLP without Catalytic Lysine
Crystals, 12, 2022
|
|
5H9A
 
 | | Crystal structure of the Apo form of human cellular retinol binding protein 1 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Retinol-binding protein 1 | | Authors: | Golczak, M, Arne, J.M, Silvaroli, J.A, Kiser, P.D, Banerjee, S. | | Deposit date: | 2015-12-26 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.381 Å) | | Cite: | Ligand Binding Induces Conformational Changes in Human Cellular Retinol-binding Protein 1 (CRBP1) Revealed by Atomic Resolution Crystal Structures.
J.Biol.Chem., 291, 2016
|
|
3RKV
 
 | | C-terminal domain of protein C56C10.10, a putative peptidylprolyl isomerase, from Caenorhabditis elegans | | Descriptor: | putative peptidylprolyl isomerase | | Authors: | Osipiuk, J, Tesar, C, Gu, M, Van Oosten-Hawle, P, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-18 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | C-terminal domain of protein C56C10.10, a putative peptidylprolyl isomerase, from Caenorhabditis elegans
To be Published
|
|
7N9O
 
 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with Aliphatic SERD S-C10(15) | | Descriptor: | 10-{[3,17beta-dihydroxyestra-1,3,5(10)-trien-7beta-yl]sulfanyl}-N-methyl-N-propyldecanamide, Estrogen receptor | | Authors: | Diennet, M, El Ezzy, M, Thiombane, K, Cotnoir-White, D, Poupart, J, Gao, Z, Mendoza, S.R, Marinier, A, Gleason, J, Mader, S.C, Fanning, S.W. | | Deposit date: | 2021-06-18 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Estrogen Receptor Alpha Ligand Binding Domain in Complex with Aliphatic SERD S-C10(15)
To Be Published
|
|
7JYP
 
 | |
7K06
 
 | |
4OJC
 
 | |
