6BOV
 
 | |
6RX3
 
 | | Crystal structure of human syncytin 2 in post-fusion conformation | | Descriptor: | CHLORIDE ION, Syncytin-2 | | Authors: | Ruigrok, K, Backovic, M, Vaney, M.C, Rey, F.A. | | Deposit date: | 2019-06-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray Structures of the Post-fusion 6-Helix Bundle of the Human Syncytins and their Functional Implications.
J.Mol.Biol., 431, 2019
|
|
6N4R
 
 | | CryoEM structure of Nav1.7 VSD2 (deactived state) in complex with the gating modifier toxin ProTx2 | | Descriptor: | Beta/omega-theraphotoxin-Tp2a, Fab heavy chain, Fab light chain, ... | | Authors: | Xu, H, Rohou, A, Arthur, C.P, Estevez, A, Ciferri, C, Payandeh, J, Koth, C.M. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin.
Cell, 176, 2019
|
|
5W9R
 
 | | Toxoplasma Gondii CDPK1 in complex with inhibitor LJQ138 | | Descriptor: | 1-tert-butyl-3-[(1H-indol-3-yl)methyl]-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1 | | Authors: | El Bakkouri, M, Lovato, D, Loppnau, P, Lin, Y.H, Rutaganaria, F, Lopez, M.S, Shokat, L, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Sibley, D, Hui, R, Walker, J.R. | | Deposit date: | 2017-06-23 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Toxoplasma Gondii CDPK1 in complex with inhibitor LJQ138
To be published
|
|
5NPH
 
 | |
5NQ2
 
 | | 'Porcine (Sus scrofa) Major Histocompatibility Complex, class I, presenting IAYERMCNI | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CALCIUM ION, ... | | Authors: | Rizkallah, P.J, Tungatt, K, Sewell, A.K. | | Deposit date: | 2017-04-19 | | Release date: | 2018-04-25 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Induction of influenza-specific local CD8 T-cells in the respiratory tract after aerosol delivery of vaccine antigen or virus in the Babraham inbred pig.
Plos Pathog., 14, 2018
|
|
5L6G
 
 | | Xylooligosaccharide oxidase from Myceliophthora thermophila C1 in complex with Xylose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FAD linked oxidase-like protein, ... | | Authors: | Rozeboom, H.J, Ferrari, A.R, Fraaije, M.W. | | Deposit date: | 2016-05-30 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of a Xylooligosaccharide Oxidase from Myceliophthora thermophila C1.
J.Biol.Chem., 291, 2016
|
|
3I4J
 
 | | Crystal structure of Aminotransferase, class III from Deinococcus radiodurans | | Descriptor: | Aminotransferase, class III, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Aminotransferase, class III from Deinococcus radiodurans
To be Published
|
|
7AML
 
 | | RET/GDNF/GFRa1 extracellular complex Cryo-EM structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha, ... | | Authors: | Adams, S.E, Earl, C.P, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2020-10-09 | | Release date: | 2021-01-13 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A two-site flexible clamp mechanism for RET-GDNF-GFR alpha 1 assembly reveals both conformational adaptation and strict geometric spacing.
Structure, 29, 2021
|
|
5FE3
 
 | | Crystal structure of human PCAF bromodomain in complex with fragment MB360 (fragment 4) | | Descriptor: | 1,2-ETHANEDIOL, 4-methoxy-1,2-benzoxazol-3-amine, Histone acetyltransferase KAT2B | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
6C7R
 
 | | BRD4 BD1 in complex with compound CF53 | | Descriptor: | Bromodomain-containing protein 4, N-(3-cyclopropyl-1-methyl-1H-pyrazol-5-yl)-7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-9H-pyrimido[4,5-b]indol-4-amine | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-01-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Discovery of CF53 as a Potent and Orally Bioavailable Bromodomain and Extra-Terminal (BET) Bromodomain Inhibitor.
J. Med. Chem., 61, 2018
|
|
5BMO
 
 | | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus | | Descriptor: | ACETATE ION, POTASSIUM ION, Putative uncharacterized protein LnmX | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Cuff, M, Endres, M, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-22 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus.
to be published
|
|
6VJG
 
 | | Csx3-I222 Crystal Form at 1.8 Angstrom Resolution | | Descriptor: | CRISPR-associated protein, Csx3 family | | Authors: | Brown, S, Charbonneau, A, Burman, N, Gauvin, C.C, Lawrence, C.M. | | Deposit date: | 2020-01-15 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Csx3 is a cyclic oligonucleotide phosphodiesterase associated with type III CRISPR-Cas that degrades the second messenger cA 4 .
J.Biol.Chem., 295, 2020
|
|
6C9V
 
 | | Mycobacterium tuberculosis adenosine kinase bound to (2R,3S,4R,5R)-2-(hydroxymethyl)-5-(6-(4-phenylpiperazin-1-yl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol | | Descriptor: | (2R,3S,4R,5R)-2-(hydroxymethyl)-5-[6-(4-phenylpiperazin-1-yl)-9H-purin-9-yl]tetrahydrofuran-3,4-diol, Adenosine kinase, GLYCEROL, ... | | Authors: | Crespo, R.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2018-01-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Drug Design of 6-Substituted Adenosine Analogues as Potent Inhibitors of Mycobacterium tuberculosis Adenosine Kinase.
J.Med.Chem., 62, 2019
|
|
5YVV
 
 | |
3RR3
 
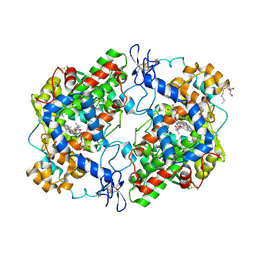 | | Structure of (R)-flurbiprofen bound to mCOX-2 | | Descriptor: | (2R)-2-(3-fluoro-4-phenyl-phenyl)propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Duggan, K.C, Hermanson, D.J, Musee, J, Prusakiewicz, J.J, Scheib, J, Carter, B.D, Banerjee, S, Oates, J.A, Marnett, L.J. | | Deposit date: | 2011-04-28 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.842 Å) | | Cite: | (R)-Profens are substrate-selective inhibitors of endocannabinoid oxygenation by COX-2.
Nat.Chem.Biol., 7, 2011
|
|
8CEB
 
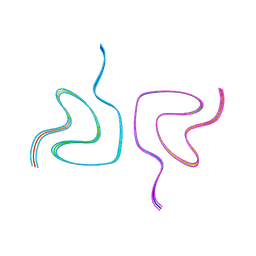 | | Type2 alpha-synuclein filament assembled in vitro by wild-type and mutant (7 residues insertion) protein | | Descriptor: | Alpha-synuclein | | Authors: | Yang, Y, Garringer, J.H, Shi, Y, Lovestam, S, Sew, P.C, Zhang, X.J, Kotecha, A, Bacioglu, M, Koto, A, Takao, M, Spillantini, G.M, Ghetti, B, Vidal, R, Murzin, G.A, Scheres, H.W.S, Goedert, M. | | Deposit date: | 2023-02-01 | | Release date: | 2023-03-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | New SNCA mutation and structures of alpha-synuclein filaments from juvenile-onset synucleinopathy.
Acta Neuropathol, 145, 2023
|
|
7S57
 
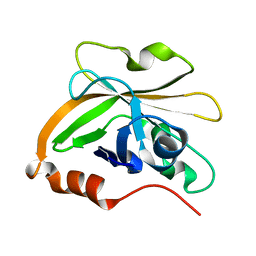 | | Structure of Sortase A from Streptococcus pyogenes with the b7-b8 loop sequence of Enterococcus faecalis Sortase A | | Descriptor: | Class A sortase, sortase A chimera | | Authors: | Svendsen, J.E, Johnson, D.A, Gao, M, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analyses of selectivity determinants in chimeric Streptococcus Class A sortase enzymes.
Protein Sci., 31, 2022
|
|
6W5E
 
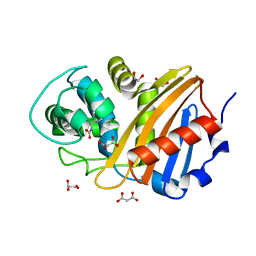 | | Class D beta-lactamase BSU-2 | | Descriptor: | 1,2-ETHANEDIOL, BSU-2 beta-lactamase, MALONATE ION | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A surface loop modulates activity of the Bacillus class D beta-lactamases.
J.Struct.Biol., 211, 2020
|
|
5II1
 
 | | Crystal Structure of the fifth bromodomain of human polybromo (PB1) in complex with 1-methylisochromeno[3,4-c]pyrazol-5(3H)-one | | Descriptor: | 1-methyl[2]benzopyrano[3,4-c]pyrazol-5(3H)-one, Protein polybromo-1 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Myrianthopoulos, V, Mikros, E, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-01 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery and Optimization of a Selective Ligand for the Switch/Sucrose Nonfermenting-Related Bromodomains of Polybromo Protein-1 by the Use of Virtual Screening and Hydration Analysis.
J.Med.Chem., 59, 2016
|
|
3RT8
 
 | |
7S53
 
 | | Structure of Sortase A from Streptococcus pyogenes with the b7-b8 loop sequence from Listeria monocytogenes Sortase A | | Descriptor: | Class A sortase, sortase A chimera | | Authors: | Johnson, D.A, Svendsen, J.E, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and biochemical analyses of selectivity determinants in chimeric Streptococcus Class A sortase enzymes.
Protein Sci., 31, 2022
|
|
8OFN
 
 | | Structure of the yellow fever virus (Asibi strain) dimeric envelope protein | | Descriptor: | Envelope glycoprotein, SULFATE ION | | Authors: | Covernton, E, Vaney, M.C, Barba-Spaeth, G, Rey, F.A. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | New insight into flavivirus maturation from structure/function studies of the yellow fever virus envelope protein complex.
Mbio, 14, 2023
|
|
1NYP
 
 | | 4th LIM domain of PINCH protein | | Descriptor: | PINCH protein, ZINC ION | | Authors: | Velyvis, A, Vaynberg, J, Vinogradova, O, Zhang, Y, Wu, C, Qin, J. | | Deposit date: | 2003-02-13 | | Release date: | 2003-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and functional insights into PINCH LIM4 domain-mediated integrin signaling
Nat.Struct.Biol., 10, 2003
|
|
4ZFJ
 
 | | Ergothioneine-biosynthetic Ntn hydrolase EgtC, apo form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Amidohydrolase EgtC | | Authors: | Vit, A, Seebeck, F.P, Blankenfeldt, W. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-01 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Ergothioneine-Biosynthesis Amidohydrolase EgtC.
Chembiochem, 16, 2015
|
|
