1THM
 
 | | CRYSTAL STRUCTURE OF THERMITASE AT 1.4 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, SODIUM ION, SULFATE ION, ... | | Authors: | Teplyakov, A.V, Kuranova, I.P, Harutyunyan, E.H. | | Deposit date: | 1992-02-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of thermitase at 1.4 A resolution.
J.Mol.Biol., 214, 1990
|
|
6W53
 
 | | Trypanosoma cruzi Malic Enzyme in complex with inhibitor (MEC070) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Malic enzyme, N-(2-chloro-5-{[4-(trifluoromethoxy)phenyl]sulfamoyl}phenyl)-3,5-difluorobenzamide | | Authors: | Mercaldi, G.F, Fagundes, M, Faria, J.N, Cordeiro, A.T. | | Deposit date: | 2020-03-12 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trypanosoma cruzi Malic Enzyme Is the Target for Sulfonamide Hits from the GSK Chagas Box.
Acs Infect Dis., 7, 2021
|
|
2J73
 
 | |
7BS8
 
 | | Bovine Pancreatic Trypsin with Benzamidine (Cryo) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Maeki, M, Ito, S, Takeda, R, Funakubo, T, Ueno, G, Ishida, A, Tani, H, Yamamoto, M, Tokeshi, M. | | Deposit date: | 2020-03-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Room-temperature crystallography using a microfluidic protein crystal array device and its application to protein-ligand complex structure analysis.
Chem Sci, 11, 2020
|
|
2VT5
 
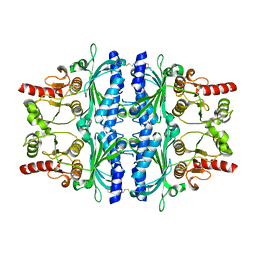 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE -1- PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH A DUAL BINDING AMP SITE INHIBITOR | | Descriptor: | 4-AMINO-N-[(2-SULFANYLETHYL)CARBAMOYL]BENZENESULFONAMIDE, FRUCTOSE-1,6-BISPHOSPHATASE 1 | | Authors: | Ruf, A, Joseph, C, Benz, J, Fol, B, Tetaz, T. | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Allosteric Fbpase Inhibitors Gain 10(5) Times in Potency When Simultaneously Binding Two Neighboring AMP Sites.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6W56
 
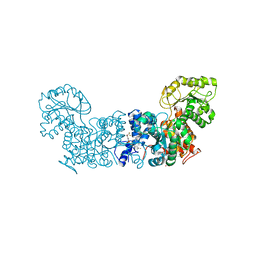 | | Trypanosoma cruzi Malic Enzyme in complex with inhibitor (MEC062) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Malic enzyme, ~{N}-[3-[[4-[bis(fluoranyl)methoxy]phenyl]sulfamoyl]phenyl]-3,5-bis(fluoranyl)benzamide | | Authors: | Mercaldi, G.F, Fagundes, M, Faria, J.N, Cordeiro, A.T. | | Deposit date: | 2020-03-12 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trypanosoma cruzi Malic Enzyme Is the Target for Sulfonamide Hits from the GSK Chagas Box.
Acs Infect Dis., 7, 2021
|
|
4IQ6
 
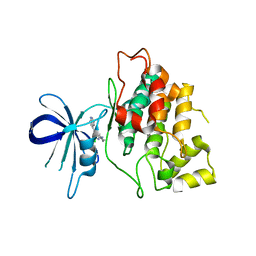 | | Gsk-3beta with inhibitor 6-chloro-N-cyclohexyl-4-(1H-pyrrolo[2,3-b]pyridin-3-yl)pyridin-2-amine | | Descriptor: | 6-chloro-N-cyclohexyl-4-(1H-pyrrolo[2,3-b]pyridin-3-yl)pyridin-2-amine, Glycogen synthase kinase-3 beta | | Authors: | Tong, Y, Stewart, K.D, Florjancic, A.S, Harlan, J.E, Merta, P.J, Przytulinska, M, Soni, N, Swinger, K.S, Zhu, H, Johnson, E.F, Shoemaker, A.R, Penning, T.D. | | Deposit date: | 2013-01-10 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Azaindole-Based Inhibitors of Cdc7 Kinase: Impact of the Pre-DFG Residue, Val 195.
ACS Med Chem Lett, 4, 2013
|
|
3ZUU
 
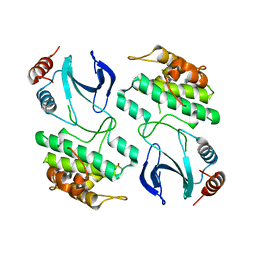 | | The structure of OST1 (D160A, S175D) kinase in complex with gold | | Descriptor: | 1,2-ETHANEDIOL, GOLD ION, Serine/threonine-protein kinase SRK2E | | Authors: | Yunta, C, Martinez-Ripoll, M, Albert, A. | | Deposit date: | 2011-07-20 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of Arabidopsis thaliana OST1 provides insights into the kinase regulation mechanism in response to osmotic stress.
J. Mol. Biol., 414, 2011
|
|
6PUL
 
 | | Structure of human MAIT A-F7 TCR in complex with human MR1 3'D-5-OP-RU | | Descriptor: | 1,3-dideoxy-1-({2,6-dioxo-5-[(E)-(2-oxopropylidene)amino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-erythro-pentitol, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Awad, W, Keller, A.N, Rossjohn, J. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The molecular basis underpinning the potency and specificity of MAIT cell antigens.
Nat.Immunol., 21, 2020
|
|
2IUO
 
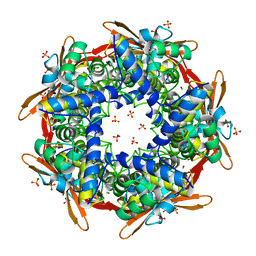 | | Site Directed Mutagenesis of Key Residues Involved in the Catalytic Mechanism of Cyanase | | Descriptor: | AZIDE ION, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Guilloton, M, Walsh, M.A, Joachimiak, A, Anderson, M.P. | | Deposit date: | 2006-06-06 | | Release date: | 2006-06-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Twin Set of Low Pka Arginines Ensures the Concerted Acid Base Catalytic Mechanism of Cyanase
To be Published
|
|
2J1W
 
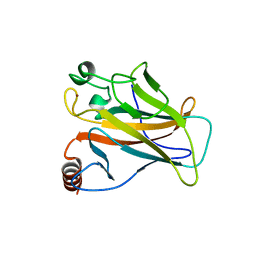 | |
1JCV
 
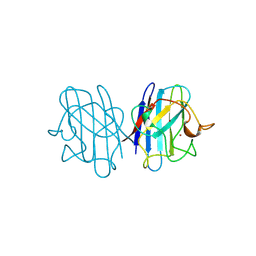 | | REDUCED BRIDGE-BROKEN YEAST CU/ZN SUPEROXIDE DISMUTASE LOW TEMPERATURE (-180C) STRUCTURE | | Descriptor: | COPPER (II) ION, CU/ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Ogihara, N.L, Parge, H.E, Hart, P.J, Weiss, M.S, Valentine, J.S, Eisenberg, D.S, Tainer, J.A. | | Deposit date: | 1995-12-07 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Unusual trigonal-planar copper configuration revealed in the atomic structure of yeast copper-zinc superoxide dismutase.
Biochemistry, 35, 1996
|
|
6PW6
 
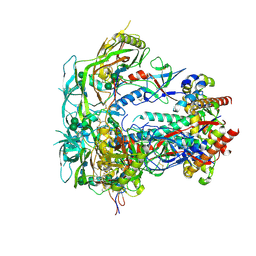 | | The HIV-1 Envelope Glycoprotein Clone BG505 SOSIP.664 in Complex with Three Copies of the Bovine Broadly Neutralizing Antibody, NC-Cow1, Fragment Antigen Binding Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Broadly Neutralizing Antibody NC-Cow1 Heavy Chain, ... | | Authors: | Berndsen, Z.T, Ward, A.B. | | Deposit date: | 2019-07-22 | | Release date: | 2020-06-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of broad HIV neutralization by a vaccine-induced cow antibody.
Sci Adv, 6, 2020
|
|
7ZY3
 
 | | Room temperature structure of Archaerhodopsin-3 obtained 110 ns after photoexcitation | | Descriptor: | Archaerhodopsin-3, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kwan, T.O.C, Judge, P.J, Moraes, I, Watts, A, Axford, D, Bada Juarez, J.F. | | Deposit date: | 2022-05-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A versatile approach to high-density microcrystals in lipidic cubic phase for room-temperature serial crystallography.
J.Appl.Crystallogr., 56, 2023
|
|
6PX2
 
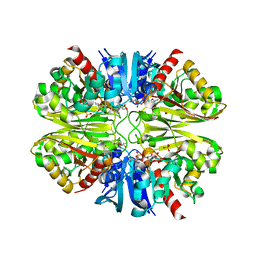 | | Acropora millepora GAPDH | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Brandt, G.S, Fields, P.A. | | Deposit date: | 2019-07-24 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Thermal stability and structure of glyceraldehyde-3-phosphate dehydrogenase from the coral Acropora millepora.
Rsc Adv, 11, 2021
|
|
5KL9
 
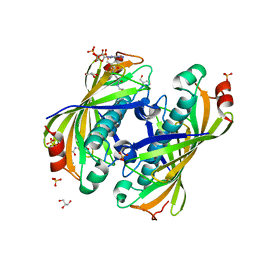 | | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with CoA | | Descriptor: | Acyl-CoA thioester hydrolase YbgC, COENZYME A, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-23 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with CoA
To Be Published
|
|
7ZTH
 
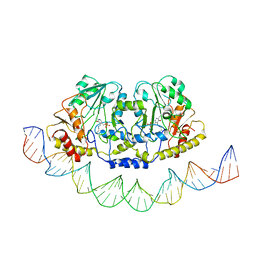 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the open conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-05-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
1K83
 
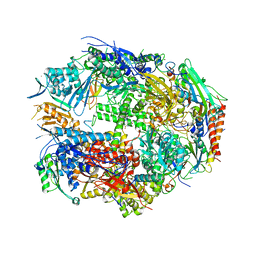 | | Crystal Structure of Yeast RNA Polymerase II Complexed with the Inhibitor Alpha Amanitin | | Descriptor: | ALPHA AMANITIN, DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.2KD POLYPEPTIDE, ... | | Authors: | Bushnell, D.A, Cramer, P, Kornberg, R.D. | | Deposit date: | 2001-10-22 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Transcription: Alpha-Amanitin-RNA Polymerase II Cocrystal at 2.8 A Resolution.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2IKG
 
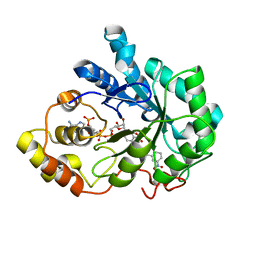 | | Aldose reductase complexed with nitrophenyl-oxadiazol type inhibitor at 1.43 A | | Descriptor: | 4-[3-(3-NITROPHENYL)-1,2,4-OXADIAZOL-5-YL]BUTANOIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2006-10-02 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural and Thermodynamic Study on Aldose Reductase: Nitro-substituted Inhibitors with Strong Enthalpic Binding Contribution
J.Mol.Biol., 368, 2007
|
|
6Q26
 
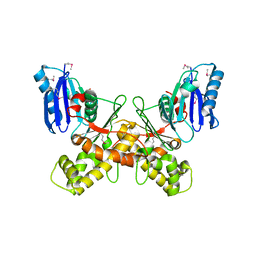 | |
6VTX
 
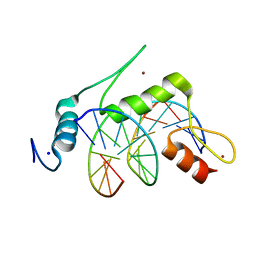 | | Crystal structure of human KLF4 zinc finger DNA binding domain in complex with NANOG DNA | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*GP*GP*TP*GP*TP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*AP*CP*AP*CP*CP*CP*CP*CP*T)-3'), Krueppel-like factor 4, ... | | Authors: | Sharma, R, Sharma, S, Choi, K.J, Ferreon, A.C.M, Ferreon, J.C, Sankaran, B, MacKenzie, K.R, Kim, C. | | Deposit date: | 2020-02-13 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Liquid condensation of reprogramming factor KLF4 with DNA provides a mechanism for chromatin organization.
Nat Commun, 12, 2021
|
|
1SYH
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND-BINDING CORE (S1S2J) IN COMPLEX WITH (S)-CPW399 AT 1.85 A RESOLUTION. | | Descriptor: | (S)-2-AMINO-3-(1,3,5,7-PENTAHYDRO-2,4-DIOXO-CYCLOPENTA[E]PYRIMIDIN-1-YL) PROIONIC ACID, Glutamate receptor 2 | | Authors: | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | Deposit date: | 2004-04-01 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
6PTB
 
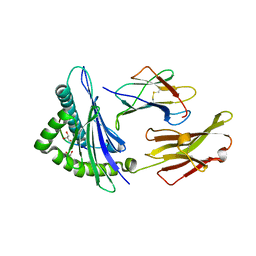 | | Crystal Structure of ILNAMIAKI peptide bound to HLA-A2 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HAUS augmin-like complex subunit 3, ... | | Authors: | Keller, G.L.J, Arbuiso, A, Baker, B.M. | | Deposit date: | 2019-07-15 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure Based Prediction of Neoantigen Immunogenicity.
Front Immunol, 10, 2019
|
|
7ZNS
 
 | |
6PTU
 
 | |
