5JRO
 
 | | The crystal structure of azoreductase from Yersinia pestis CO92 in its Apo form | | Descriptor: | FMN-dependent NADH-azoreductase, GLYCEROL | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-06 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The crystal structure of azoreductase from Yersinia pestis CO92 in its Apo form
To Be Published
|
|
6PH1
 
 | | T4 lysozyme pseudo-wild type soaked in TEMPOL | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Endolysin, ... | | Authors: | Cuneo, M.J, Myles, D.A, Li, L. | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.632 Å) | | Cite: | Making hydrogens stand out: Enhanced neutron diffraction from biological crystals using dynamic nuclear polarization
To be published
|
|
8SLA
 
 | | Cryo-EM structure of the rat TRPM5 channel in trace calcium, trace-2 | | Descriptor: | Transient receptor potential cation channel subfamily M member 5 | | Authors: | Karuppan, S, Schrag, L.G, Jara-Oseguera, A, Zubcevic, L. | | Deposit date: | 2023-04-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural dynamics at cytosolic interprotomer interfaces control gating of a mammalian TRPM5 channel.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7L5D
 
 | | The crystal structure of SARS-CoV-2 Main Protease in complex with demethylated analog of masitinib | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tan, K, Maltseva, N.I, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-21 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2.
Science, 373, 2021
|
|
6RRO
 
 | |
6RRL
 
 | |
5MQL
 
 | | Crystal structure of dCK mutant C3S in complex with masitinib and UDP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, Deoxycytidine kinase, MAGNESIUM ION, ... | | Authors: | Rebuffet, E, Hammam, K, Saez-Ayala, M, Gros, L, Lopez, S, Hajem, B, Humbert, M, Baudelet, E, Audebert, S, Betzi, S, Lugari, A, Combes, S, Pez, D, Letard, S, Mansfield, C, Moussy, A, de Sepulveda, P, Morelli, X, Dubreuil, P. | | Deposit date: | 2016-12-20 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Dual protein kinase and nucleoside kinase modulators for rationally designed polypharmacology.
Nat Commun, 8, 2017
|
|
6F1U
 
 | | N terminal region of dynein tail domains in complex with dynactin filament and BICDR-1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARP1 actin related protein 1 homolog A, BICD family-like cargo adapter 1, ... | | Authors: | Urnavicius, L, Lau, C.K, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-11-23 | | Release date: | 2018-01-17 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
4XMV
 
 | |
6RSM
 
 | | Solution NMR structure of the peptide 12530 from medicinal leech Hirudo medicinalis in dodecylphosphocholine micelles | | Descriptor: | peptide 12530 | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Grafskaia, E.N, Arseniev, A.S, Lazarev, V.N. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Medicinal leech antimicrobial peptides lacking toxicity represent a promising alternative strategy to combat antibiotic-resistant pathogens.
Eur.J.Med.Chem., 180, 2019
|
|
6F38
 
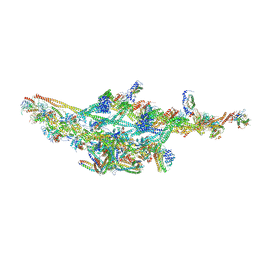 | | Cryo-EM structure of two dynein tail domains bound to dynactin and HOOK3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | Authors: | Lau, C.K, Urnavicius, L, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-11-28 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
7Z14
 
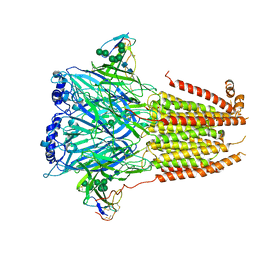 | | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with a short-chain neurotoxin. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine receptor subunit alpha, ... | | Authors: | Nys, M.A.E.M, Zarkadas, E, Ulens, C, Nury, H. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-17 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | The molecular mechanism of snake short-chain alpha-neurotoxin binding to muscle-type nicotinic acetylcholine receptors.
Nat Commun, 13, 2022
|
|
7KKL
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody mNb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
8SL8
 
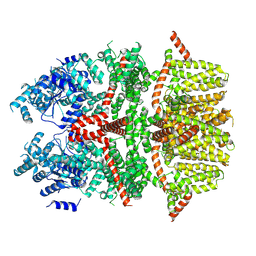 | | Cryo-EM structure of the rat TRPM5 channel in trace calcium, trace-1 | | Descriptor: | Transient receptor potential cation channel subfamily M member 5 | | Authors: | Karuppan, S, Schrag, L.G, Jara-Oseguera, A, Zubcevic, L. | | Deposit date: | 2023-04-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural dynamics at cytosolic interprotomer interfaces control gating of a mammalian TRPM5 channel.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6V8O
 
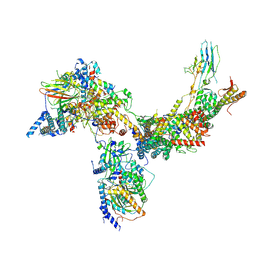 | | RSC core | | Descriptor: | Chromatin structure-remodeling complex protein RSC14, Chromatin structure-remodeling complex protein RSC3, Chromatin structure-remodeling complex protein RSC30, ... | | Authors: | Patel, A.B, Moore, C.M, Greber, B.J, Nogales, E. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement.
Elife, 8, 2019
|
|
5K79
 
 | | Structure and anti-HIV activity of CYT-CVNH, a new cyanovirin-n homolog | | Descriptor: | 1,2-ETHANEDIOL, Cyanovirin-N domain protein, DI(HYDROXYETHYL)ETHER | | Authors: | Matei, E, Basu, R, Furey, W, Shi, J, Calnan, C, Aiken, C, Gronenborn, A.M. | | Deposit date: | 2016-05-25 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Glycan Binding of a New Cyanovirin-N Homolog.
J.Biol.Chem., 291, 2016
|
|
6F58
 
 | | Crystal structure of human Brachyury (T) in complex with DNA | | Descriptor: | Brachyury protein, DNA (5'-D(*AP*AP*TP*TP*TP*CP*AP*CP*AP*CP*CP*TP*AP*GP*GP*TP*GP*TP*GP*AP*AP*AP*TP*T)-3'), SODIUM ION | | Authors: | Newman, J.A, Gavard, A.E, Krojer, T, Shrestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-12-01 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Crystal structure of human Brachyury (T) in complex with DNA
To Be Published
|
|
6S98
 
 | |
6S9N
 
 | | Designed Armadillo Repeat protein Lock2 fused to target peptide KRKRKAKLSF | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Lock2_KRKRKAKLSF | | Authors: | Ernst, P, Zosel, F, Reichen, C, Schuler, B, Pluckthun, A. | | Deposit date: | 2019-07-15 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design of a Peptide Lock for Modular Peptide Binders.
Acs Chem.Biol., 15, 2020
|
|
6VBM
 
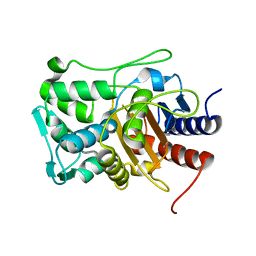 | | Crystal structure of a S310A mutant of PBP2 from Neisseria gonorrhoeae | | Descriptor: | PHOSPHATE ION, Probable peptidoglycan D,D-transpeptidase PenA | | Authors: | Singh, A, Davies, C. | | Deposit date: | 2019-12-19 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Mutations in Neisseria gonorrhoeae penicillin-binding protein 2 associated with extended-spectrum cephalosporin resistance create an energetic barrier against acylation via restriction of protein dynamics
J.Biol.Chem., 2020
|
|
6SBO
 
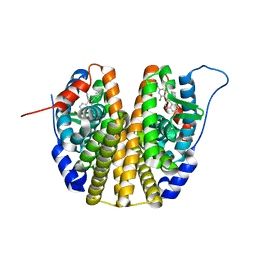 | | Estrogen receptor mutant L536S | | Descriptor: | 6-(2,4-dichlorophenyl)-5-[4-[(3~{S})-1-(3-fluoranylpropyl)pyrrolidin-3-yl]oxyphenyl]-8,9-dihydro-7~{H}-benzo[7]annulene-2-carboxylic acid, Estrogen receptor | | Authors: | Vallee, F, Steier, V, Rak, A. | | Deposit date: | 2019-07-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Discovery of 6-(2,4-Dichlorophenyl)-5-[4-[(3S)-1-(3-fluoropropyl)pyrrolidin-3-yl]oxyphenyl]-8,9-dihydro-7H-benzo[7]annulene-2-carboxylic acid (SAR439859), a Potent and Selective Estrogen Receptor Degrader (SERD) for the Treatment of Estrogen-Receptor-Positive Breast Cancer.
J.Med.Chem., 63, 2020
|
|
6F5Q
 
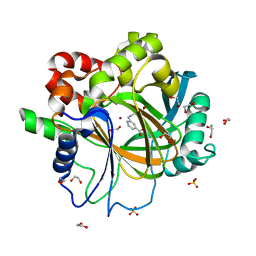 | | Crystal Structure of KDM4D with GF026 ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(methylsulfonylamino)ethylamino]pyridine-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-12-02 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal Structure of KDM4D with GF026 ligand
To be published
|
|
5JY8
 
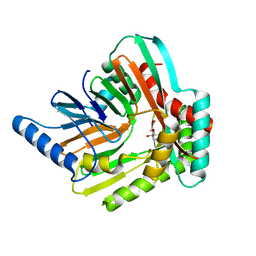 | | An iron-bound structure of the isochorismate synthase EntC | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, FE (III) ION, Isochorismate synthase EntC | | Authors: | Meneely, K.M, Sundlov, J.A, Gulick, A.M, Lamb, A.L. | | Deposit date: | 2016-05-13 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.942 Å) | | Cite: | An Open and Shut Case: The Interaction of Magnesium with MST Enzymes.
J.Am.Chem.Soc., 138, 2016
|
|
8KB6
 
 | | Crystal Structure of Canine TNF-alpha | | Descriptor: | Tumor necrosis factor | | Authors: | Lee, C.C, Wang, A.H.-J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.850166 Å) | | Cite: | Structure-based development of a canine TNF-alpha-specific antibody using adalimumab as a template.
Protein Sci., 33, 2024
|
|
4XQU
 
 | | Crystal structure of hemagglutinin from Jiangxi-Donghu (2013) H10N8 influenza virus in complex with 3'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Tzarum, N, Zhang, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2015-01-20 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | A Human-Infecting H10N8 Influenza Virus Retains a Strong Preference for Avian-type Receptors.
Cell Host Microbe, 17, 2015
|
|
