5F0J
 
 | | Structure of retromer VPS26-VPS35 subunits bound to SNX3 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Lucas, M, Gershlick, D, Vidaurrazaga, A, Rojas, A.L, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Mechanism for Cargo Recognition by the Retromer Complex.
Cell, 167, 2016
|
|
6T2M
 
 | | VDR-ZK168281 complex | | Descriptor: | Nuclear receptor coactivator 1, Vitamin D3 receptor A, ethyl (~{Z})-3-[1-[(~{E},1~{R},4~{R})-4-[(1~{R},3~{a}~{S},4~{E},7~{a}~{R})-7~{a}-methyl-4-[(2~{Z})-2-[(3~{S},5~{R})-2-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]-1-oxidanyl-pent-2-enyl]cyclopropyl]prop-2-enoate | | Authors: | Rochel, N, Belorusova, A.Y. | | Deposit date: | 2019-10-09 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Analysis of VDR Complex with ZK168281 Antagonist.
J.Med.Chem., 63, 2020
|
|
6YNI
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_036 | | Descriptor: | (2~{R},3~{R})-2-(3-chlorophenyl)-3,5,5-trimethyl-morpholine, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
3N3H
 
 | | Erythrina corallodendron lectin mutant (Y106G) in complex with citrate | | Descriptor: | CALCIUM ION, CITRIC ACID, Lectin, ... | | Authors: | Thamotharan, S, Karthikeyan, T, Kulkarni, K.A, Shetty, K.N, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2010-05-20 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modification of the sugar specificity of a plant lectin: structural studies on a point mutant of Erythrina corallodendron lectin.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5F1R
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (C10) | | Descriptor: | (3~{R})-8-cyclopropyl-7-(naphthalen-1-ylmethyl)-5-oxidanylidene-2,3-dihydro-[1,3]thiazolo[3,2-a]pyridine-3-carboxylic acid, Listeriolysin regulatory protein | | Authors: | Begum, A, Grundstrom, C, Good, J.A.D, Andersson, C, ALmqvist, F, Johansson, J, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Attenuating Listeria monocytogenes Virulence by Targeting the Regulatory Protein PrfA.
Cell Chem Biol, 23, 2016
|
|
6YPX
 
 | | Human histidine triad nucleotide-binding protein 2 (hHINT2) refined to 2.11 A in C2221 space group | | Descriptor: | CACODYLATE ION, GLYCEROL, Histidine triad nucleotide-binding protein 2, ... | | Authors: | Dolot, R.D, Wlodarczyk, A, Bujacz, G.D, Nawrot, B.C. | | Deposit date: | 2020-04-16 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Biochemical, crystallographic and biophysical characterization of histidine triad nucleotide-binding protein 2 with different ligands including a non-hydrolyzable analog of Ap4A.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
7BUZ
 
 | |
6YRO
 
 | | Streptococcus suis SadP mutant - N285D | | Descriptor: | GLYCEROL, SODIUM ION, SadP | | Authors: | Papageorgiou, A.C, Haataja, S. | | Deposit date: | 2020-04-20 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The binding mechanism of the virulence factor Streptococcus suis adhesin P subtype to globotetraosylceramide is associated with systemic disease.
J.Biol.Chem., 295, 2020
|
|
4YZ2
 
 | | Crystal Structure of Streptococcus pneumoniae NanC, in complex with 2-deoxy-2,3-didehydro-N-acetylneuraminic acid. | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, SULFATE ION, Sialidase NanC | | Authors: | Lukacik, P, Owen, C.D, Potter, J.A, Taylor, G.L, Walsh, M.A. | | Deposit date: | 2015-03-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
7OBQ
 
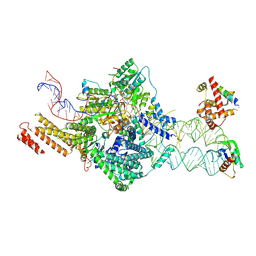 | | SRP-SR at the distal site conformation | | Descriptor: | EM14S01-3B_G0054400.mRNA.1.CDS.1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jomaa, A, Ban, N. | | Deposit date: | 2021-04-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular mechanism of cargo recognition and handover by the mammalian signal recognition particle.
Cell Rep, 36, 2021
|
|
5NZR
 
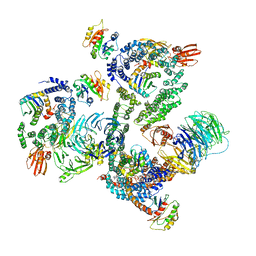 | | The structure of the COPI coat leaf | | Descriptor: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
6C65
 
 | | Crystal Structure of the Mango-II-A22U Fluorescent Aptamer Bound to TO1-Biotin | | Descriptor: | 4-[(3-{2-[(2-methoxyethyl)amino]-2-oxoethyl}-1,3-benzothiazol-3-ium-2-yl)methyl]-1-methylquinolin-1-ium, POTASSIUM ION, RNA (35-MER), ... | | Authors: | Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2018-01-17 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.80005169 Å) | | Cite: | Crystal Structures of the Mango-II RNA Aptamer Reveal Heterogeneous Fluorophore Binding and Guide Engineering of Variants with Improved Selectivity and Brightness.
Biochemistry, 57, 2018
|
|
4YZV
 
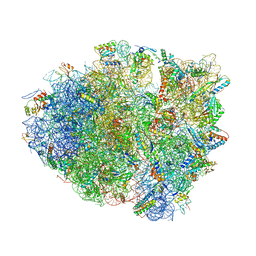 | | Precleavage 70S structure of the P. vulgaris HigB deltaH92 toxin bound to the ACA codon | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Schureck, M.A, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2015-03-25 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Defining the mRNA recognition signature of a bacterial toxin protein.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5F53
 
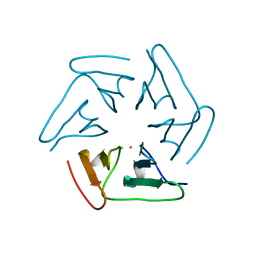 | |
6T61
 
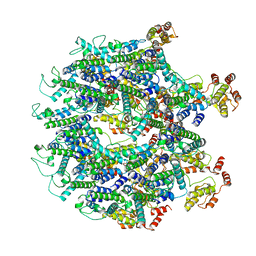 | | A model of the EIAV CA-SP hexamer (C2) from Gag-deltaMA tubes assembled at pH8 | | Descriptor: | Gag polyprotein | | Authors: | Dick, R.A, Xu, C, Morado, D.R, Kravchuk, V, Ricana, C.L, Lyddon, T.D, Broad, A.M, Feathers, J.R, Johnson, M.C, Vogt, V.M, Perilla, J.R, Briggs, J.A.G, Schur, F.K.M. | | Deposit date: | 2019-10-17 | | Release date: | 2020-01-15 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly.
Plos Pathog., 16, 2020
|
|
6FY6
 
 | | Concerted dynamics of metallo-base pairs in an A/B-form helical transition (major species) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*TP*CP*AP*TP*GP*AP*TP*AP*CP*G)-3')_major, MERCURY (II) ION | | Authors: | Schmidt, O.P, Jurt, S, Johannsen, S, Karimi, A, Sigel, R.K.O, Luedtke, N.W. | | Deposit date: | 2018-03-11 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Concerted dynamics of metallo-base pairs in an A/B-form helical transition.
Nat Commun, 10, 2019
|
|
6C6S
 
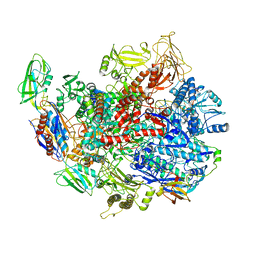 | | CryoEM structure of E.coli RNA polymerase elongation complex bound with RfaH | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Artsimovitch, I, Landick, R, Darst, S.A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators.
Cell, 173, 2018
|
|
6N6X
 
 | |
4ZE3
 
 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) Y140H mutant complexed with fluconazole | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sagatova, A, Keniya, M.V, Wilson, R, Monk, B.C, Tyndall, J.D.A. | | Deposit date: | 2015-04-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Triazole resistance mediated by mutations of a conserved active site tyrosine in fungal lanosterol 14 alpha-demethylase.
Sci Rep, 6, 2016
|
|
7OBR
 
 | | RNC-SRP early complex | | Descriptor: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Ban, N. | | Deposit date: | 2021-04-23 | | Release date: | 2021-07-21 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular mechanism of cargo recognition and handover by the mammalian signal recognition particle.
Cell Rep, 36, 2021
|
|
7T75
 
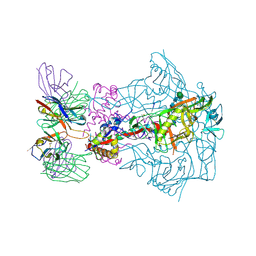 | | HIV-1 Envelope ApexGT2 in complex with RM20A3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV Envelope ApexGT2 gp120, ... | | Authors: | Berndsen, Z.T, Ward, A.B. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-28 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Human immunoglobulin repertoire analysis guides design of vaccine priming immunogens targeting HIV V2-apex broadly neutralizing antibody precursors.
Immunity, 55, 2022
|
|
6X05
 
 | |
7SMI
 
 | | Crystal Structure of L-galactose dehydrogenase from Spinacia oleracea | | Descriptor: | L-galactose dehydrogenase | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2021-10-26 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Characterization of L-Galactose Dehydrogenase: An Essential Enzyme for Vitamin C Biosynthesis.
Plant Cell.Physiol., 63, 2022
|
|
6TQS
 
 | | The crystal structure of the MSP domain of human MOSPD2 in complex with the conventional FFAT motif of ORP1. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
4U0U
 
 | | Wild type eukaryotic fic domain containing protein with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, MAGNESIUM ION, ... | | Authors: | Cole, A.R, Bunney, T.D, Katan, M. | | Deposit date: | 2014-07-14 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Crystal structure of the human, FIC-domain containing protein HYPE and implications for its functions.
Structure, 22, 2014
|
|
