6RWN
 
 | | SIVrcm intasome in complex with dolutegravir | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6RTY
 
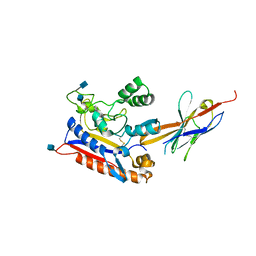 | | Crystal structure of the Patched ectodomain in complex with nanobody NB64 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Llama-derived nanobody NB64, Protein patched homolog 1 | | Authors: | Rudolf, A.F, Kowatsch, C, El Omari, K, Malinauskas, T, Kinnebrew, M, Ansell, T.B, Bishop, B, Pardon, E, Schwab, R.A, Qian, M, Duman, R, Covey, D.F, Steyaert, J, Wagner, A, Sansom, M.S.P, Rohatgi, R, Siebold, C. | | Deposit date: | 2019-05-27 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Nat.Chem.Biol., 15, 2019
|
|
6RX1
 
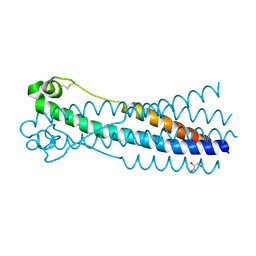 | | Crystal structure of human syncytin 1 in post-fusion conformation | | Descriptor: | CHLORIDE ION, GLYCEROL, Syncytin-1 | | Authors: | Ruigrok, K, Backovic, M, Vaney, M.C, Rey, F.A. | | Deposit date: | 2019-06-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structures of the Post-fusion 6-Helix Bundle of the Human Syncytins and their Functional Implications.
J.Mol.Biol., 431, 2019
|
|
4R2X
 
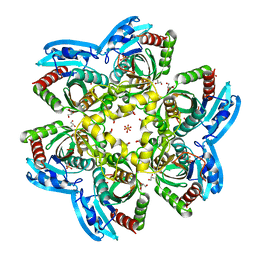 | | Unique conformation of uridine and asymmetry of the hexameric molecule revealed in the high-resolution structures of Shewanella oneidensis uridine phosphorylase in the free form and in complex with uridine | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Safonova, T.N, Mordkovich, N.N, Manuvera, V.A, Veiko, V.P, Popov, V.O, Polyakov, K.M. | | Deposit date: | 2014-08-13 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | High-syn conformation of uridine and asymmetry of the hexameric molecule revealed in the high-resolution structures of Shewanella oneidensis MR-1 uridine phosphorylase in the free form and in complex with uridine.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8EAT
 
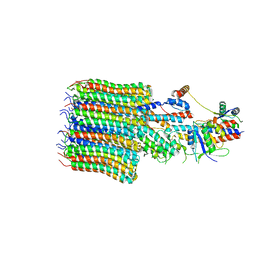 | | Yeast VO missing subunits a, e, and f in complex with Vma12-22p | | Descriptor: | V-type proton ATPase subunit F, V-type proton ATPase subunit c, V-type proton ATPase subunit c', ... | | Authors: | Wang, H, Bueler, S.A, Rubinstein, J.L. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of V-ATPase V O region assembly by Vma12p, 21p, and 22p.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8TRG
 
 | |
5X5A
 
 | | Human thymidylate synthase bound with phosphate ion | | Descriptor: | PHOSPHATE ION, Thymidylate synthase | | Authors: | Chen, D, Jansson, A, Larsson, A, Nordlund, P. | | Deposit date: | 2017-02-15 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural analyses of human thymidylate synthase reveal a site that may control conformational switching between active and inactive states
J. Biol. Chem., 292, 2017
|
|
8DZG
 
 | | Cryo-EM structure of bundle-forming pilus extension ATPase from E.coli in the presence of ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BfpD, MAGNESIUM ION, ... | | Authors: | Nayak, A.R, Zhao, J, Donnenberg, M.S, Samso, M. | | Deposit date: | 2022-08-07 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM Structure of the Type IV Pilus Extension ATPase from Enteropathogenic Escherichia coli.
Mbio, 13, 2022
|
|
5ML8
 
 | | The crystal structure of PDE6D in complex to inhibitor-4 | | Descriptor: | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta, ~{N}4-[(4-chlorophenyl)methyl]-~{N}4-cyclopentyl-~{N}1-(phenylmethyl)-~{N}1-(piperidin-4-ylmethyl)benzene-1,4-disulfonamide | | Authors: | Fansa, E.K, Martin-Gago, P, Waldmann, H, Wittinghofer, A. | | Deposit date: | 2016-12-06 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A PDE6 delta-KRas Inhibitor Chemotype with up to Seven H-Bonds and Picomolar Affinity that Prevents Efficient Inhibitor Release by Arl2.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5K09
 
 | | Crystal Structure of COMT in complex with a thiazole ligand | | Descriptor: | 5-{3-[(4-methoxyphenyl)methyl]-1H-pyrazol-5-yl}-2,4-dimethyl-1,3-thiazole, Catechol O-methyltransferase, PHOSPHATE ION, ... | | Authors: | Ehler, A, Rodriguez-Sarmiento, R.M, Rudolph, M.G. | | Deposit date: | 2016-05-17 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of Potent and Druglike Nonphenolic Inhibitors for Catechol O-Methyltransferase Derived from a Fragment Screening Approach Targeting the S-Adenosyl-l-methionine Pocket.
J. Med. Chem., 59, 2016
|
|
7RFB
 
 | | Crystal structure of broadly neutralizing antibody mAb1198 in complex with Hepatitis C virus envelope glycoprotein E2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Flyak, A.I, Bjorkman, P.J. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Analysis of antibodies from HCV elite neutralizers identifies genetic determinants of broad neutralization.
Immunity, 55, 2022
|
|
6RVX
 
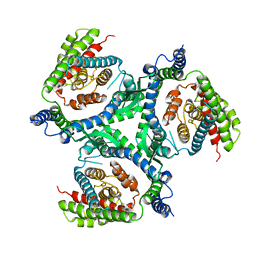 | |
6U5J
 
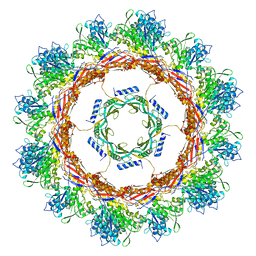 | | CryoEM Structure of Pyocin R2 - postcontracted - collar | | Descriptor: | Collar PA0615, Sheath PA0622 | | Authors: | Ge, P, Avaylon, J, Scholl, D, Shneider, M.M, Browning, C, Buth, S.A, Plattner, M, Ding, K, Leiman, P.G, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Action of a minimal contractile bactericidal nanomachine.
Nature, 580, 2020
|
|
5K39
 
 | | THE TYPE II COHESIN DOCKERIN COMPLEX FROM CLOSTRIDIUM THERMOCELLUM | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Dockerin module from a protein of unknown function | | Authors: | Viegas, A, Pinheiro, B, Bras, J.L.A, Romao, M.J, Alves, V, Carvalho, A.L, Fontes, C.M.G.A. | | Deposit date: | 2016-05-19 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Diverse specificity of cellulosome attachment to the bacterial cell surface.
Sci Rep, 6, 2016
|
|
6UHA
 
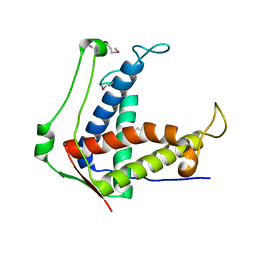 | |
5H2M
 
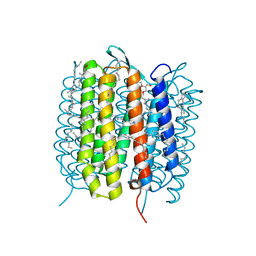 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 13.8 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
6RYZ
 
 | | SalL with S-adenosyl methionine | | Descriptor: | 1,2-ETHANEDIOL, Adenosyl-chloride synthase, CHLORIDE ION, ... | | Authors: | McKean, I, Frese, A, Cuetos, A, Burley, G, Grogan, G. | | Deposit date: | 2019-06-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | S-Adenosyl Methionine Cofactor Modifications Enhance the Biocatalytic Repertoire of Small Molecule C-Alkylation.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6S21
 
 | | Metabolism of multiple glycosaminoglycans by bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus (BT33494S-sulf) | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, CALCIUM ION, Endo-4-O-sulfatase | | Authors: | Ndeh, D, Basle, A, Strahl, H, Henrissat, B, Terrapon, N, Cartmell, A. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Metabolism of multiple glycosaminoglycans by Bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus.
Nat Commun, 11, 2020
|
|
5KGV
 
 | |
6UKS
 
 | | ATPgammaS bound mBcs1 | | Descriptor: | MAGNESIUM ION, Mitochondrial chaperone BCS1, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Tang, W.K, Borgnia, M.J, Hsu, A.L, Xia, D. | | Deposit date: | 2019-10-05 | | Release date: | 2020-02-05 | | Last modified: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of AAA protein translocase Bcs1 suggest translocation mechanism of a folded protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7RFC
 
 | |
8TTO
 
 | |
8U7I
 
 | | Structure of the phage immune evasion protein Gad1 bound to the Gabija GajAB complex | | Descriptor: | Endonuclease GajA, Gabija Anti-Defense 1, Gabija protein GajB | | Authors: | Antine, S.P, Johnson, A.G, Mooney, S.E, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2023-09-15 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural basis of Gabija anti-phage defence and viral immune evasion.
Nature, 625, 2024
|
|
4QSA
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 2-chloro-4-{[(4-methyl-6-oxo-1,6-dihydropyrimidin-2-yl)thio]acetyl}benzenesulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-4-{[(4-methyl-6-oxo-1,6-dihydropyrimidin-2-yl)sulfanyl]acetyl}benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-07-03 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Intrinsic Thermodynamics and Structure Correlation of Benzenesulfonamides with a Pyrimidine Moiety Binding to Carbonic Anhydrases I, II, VII, XII, and XIII
Plos One, 9, 2014
|
|
8DVB
 
 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with (1'-(4-((1-butylpyrrolidin-3-yl)methoxy)phenyl)-6'-hydroxy-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl)(phenyl)methanone | | Descriptor: | Estrogen receptor, [(1'R)-1'-(4-{[(3R)-1-(3-fluoropropyl)pyrrolidin-3-yl]methoxy}phenyl)-6'-hydroxy-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl](phenyl)methanone | | Authors: | Hancock, G.R, Young, K.S, Hosfield, D.J, Joiner, C, Sullivan, E.A, Yildz, Y, Laine, M, Greene, G.L, Fanning, S.W. | | Deposit date: | 2022-07-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Unconventional isoquinoline-based SERMs elicit fulvestrant-like transcriptional programs in ER+ breast cancer cells.
NPJ Breast Cancer, 8, 2022
|
|
