8RIH
 
 | | Crystal structure of the Saccharomyces cerevisiae URH1p riboside hydrolase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Uridine ribohydrolase | | Authors: | Degano, M, Carriles, A.A. | | Deposit date: | 2023-12-18 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure-Function Insights into the Dual Role in Nucleobase and Nicotinamide Metabolism and a Possible Use in Cancer Gene Therapy of the URH1p Riboside Hydrolase.
Int J Mol Sci, 25, 2024
|
|
6ME8
 
 | | XFEL crystal structure of human melatonin receptor MT2 (N86D) in complex with 2-phenylmelatonin | | Descriptor: | N-[2-(5-methoxy-2-phenyl-1H-indol-3-yl)ethyl]acetamide, Soluble cytochrome b562,Melatonin receptor type 1B,Rubredoxin, ZINC ION | | Authors: | Johansson, L.C, Stauch, B, McCorvy, J, Han, G.W, Patel, N, Batyuk, A, Gati, C, Li, C, Grandner, J, Hao, S, Olsen, R.H.J, Tribo, A.R, Zaare, S, Zhu, L, Zatsepin, N.A, Weierstall, U, Liu, W, Roth, B.L, Katritch, V, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity.
Nature, 569, 2019
|
|
1ASP
 
 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, COPPER (II) ION, ... | | Authors: | Messerschmidt, A, Luecke, H, Huber, R. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
8BKF
 
 | | Structure of E. coli Class 2 L-asparaginase EcAIII, mutant M200T (crystal M200T#o) | | Descriptor: | CHLORIDE ION, Isoaspartyl peptidase subunit alpha, Isoaspartyl peptidase subunit beta, ... | | Authors: | Sciuk, A, Ruszkowski, M, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-11-09 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.221 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
5MQT
 
 | | Crystal structure of dCK mutant C3S in complex with imatinib and UDP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Deoxycytidine kinase, ... | | Authors: | Saez-Ayala, M, Rebuffet, E, Hammam, K, Gros, L, Lopez, S, Hajem, B, Humbert, M, Baudelet, E, Audebert, S, Betzi, S, Lugari, A, Combes, S, Pez, D, Letard, S, Mansfield, C, Moussy, A, de Sepulveda, P, Morelli, X, Dubreuil, P. | | Deposit date: | 2016-12-20 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dual protein kinase and nucleoside kinase modulators for rationally designed polypharmacology.
Nat Commun, 8, 2017
|
|
7RU6
 
 | |
4R7R
 
 | | Crystal Structure of Putative Lipoprotein from Clostridium perfringens | | Descriptor: | GLYCEROL, Putative lipoprotein | | Authors: | Kim, Y, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Crystal Structure of Putative Lipoprotein from Clostridium perfringens
To be Published
|
|
4Y3M
 
 | |
6POJ
 
 | |
8BP9
 
 | | Structure of E. coli Class 2 L-asparaginase EcAIII, mutant M200W (crystal M200W#2) | | Descriptor: | CHLORIDE ION, Isoaspartyl peptidase subunit alpha, Isoaspartyl peptidase subunit beta, ... | | Authors: | Sciuk, A, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
4Y54
 
 | | Endothiapepsin in complex with fragment 56 | | Descriptor: | (2R)-(3-chlorophenyl)(hydroxy)ethanoic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Radeva, N, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
6F9G
 
 | | Ligand binding domain of P. putida KT2440 polyamine chemorecpetors McpU in complex putrescine. | | Descriptor: | 1,4-DIAMINOBUTANE, ACETATE ION, GLYCEROL, ... | | Authors: | Gavira, J.A, Conejero-Muriel, M.T, Ortega, A, Martin-Mora, D, Corral-Lugo, A, Morel, B, Krell, T. | | Deposit date: | 2017-12-14 | | Release date: | 2018-03-28 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Structural Basis for Polyamine Binding at the dCACHE Domain of the McpU Chemoreceptor from Pseudomonas putida.
J. Mol. Biol., 430, 2018
|
|
6SGU
 
 | | Cryo-EM structure of Escherichia coli AcrB and DARPin in Saposin A-nanodisc | | Descriptor: | DARPin, Multidrug efflux pump subunit AcrB | | Authors: | Szewczak-Harris, A, Du, D, Newman, C, Neuberger, A, Luisi, B.F. | | Deposit date: | 2019-08-05 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Interactions of a Bacterial RND Transporter with a Transmembrane Small Protein in a Lipid Environment.
Structure, 28, 2020
|
|
7JYE
 
 | | Human Liver Receptor Homolog-1 in Complex with 9ChoP and a Fragment of Tif2 | | Descriptor: | 9-[(3~{a}~{R},6~{R},6~{a}~{R})-6-oxidanyl-3-phenyl-3~{a}-(1-phenylethenyl)-4,5,6,6~{a}-tetrahydro-1~{H}-pentalen-2-yl]nonyl 2-(trimethyl-$l^{4}-azanyl)ethyl hydrogen phosphate, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | D'Agostino, E.H, Mays, S.G, Ortlund, E.A. | | Deposit date: | 2020-08-30 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Tapping into a phospholipid-LRH-1 axis yields a powerful anti-inflammatory agent with in vivo activity against colitis
To Be Published
|
|
6SNZ
 
 | | Crystal structure of lamin A coil1b tetramer | | Descriptor: | GLYCEROL, Prelamin-A/C | | Authors: | Lilina, A.V, Chernyatina, A.A, Guzenko, D, Strelkov, S.V. | | Deposit date: | 2019-08-28 | | Release date: | 2019-10-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Lateral A11type tetramerization in lamins.
J.Struct.Biol., 209, 2020
|
|
1BCF
 
 | | THE STRUCTURE OF A UNIQUE, TWO-FOLD SYMMETRIC, HAEM-BINDING SITE | | Descriptor: | BACTERIOFERRITIN, MANGANESE (II) ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Frolow, F, Kalb(Gilboa), A.J, Yariv, J. | | Deposit date: | 1993-12-06 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a unique twofold symmetric haem-binding site.
Nat.Struct.Biol., 1, 1994
|
|
6MFV
 
 | | Crystal structure of the Signal Transduction ATPase with Numerous Domains (STAND) protein with a tetratricopeptide repeat sensor PH0952 from Pyrococcus horikoshii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, tetratricopeptide repeat sensor PH0952 | | Authors: | Lisa, M.N, Alzari, P.M, Haouz, A, Danot, O. | | Deposit date: | 2018-09-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Double autoinhibition mechanism of signal transduction ATPases with numerous domains (STAND) with a tetratricopeptide repeat sensor.
Nucleic Acids Res., 47, 2019
|
|
6PQK
 
 | |
3LY5
 
 | | DDX18 dead-domain | | Descriptor: | ATP-dependent RNA helicase DDX18, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Schutz, P, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Kotzsch, A, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-26 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
7RUG
 
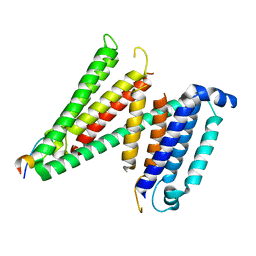 | | Human SERINC3-DeltaICL4 | | Descriptor: | Serine incorporator 3, SiA | | Authors: | Purdy, M.D, Leonhardt, S.A, Yeager, M. | | Deposit date: | 2021-08-17 | | Release date: | 2022-08-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Antiviral HIV-1 SERINC restriction factors disrupt virus membrane asymmetry.
Nat Commun, 14, 2023
|
|
6SAA
 
 | | M-TRTX-Preg1a (Poecilotheria regalis) | | Descriptor: | U1-theraphotoxin-Pf3 | | Authors: | Meudal, H, Landon, C, Delmas, A.F. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2020-08-26 | | Method: | SOLUTION NMR | | Cite: | A Venomics Approach Coupled to High-Throughput Toxin Production Strategies Identifies the First Venom-Derived Melanocortin Receptor Agonists.
J.Med.Chem., 63, 2020
|
|
6SAU
 
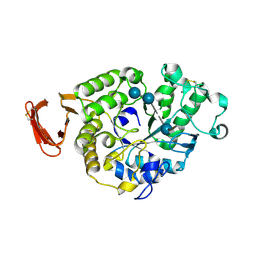 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance. | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, SODIUM ION, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tinaqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
8W01
 
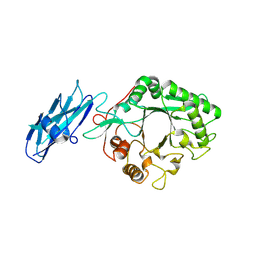 | |
8BQO
 
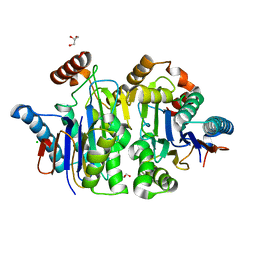 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant M200I | | Descriptor: | CHLORIDE ION, GLYCEROL, Isoaspartyl peptidase subunit alpha, ... | | Authors: | Sciuk, A, Ruszkowski, M, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-11-21 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
4RCI
 
 | | Crystal structure of YTHDF1 YTH domain | | Descriptor: | UNKNOWN ATOM OR ION, YTH domain-containing family protein 1 | | Authors: | Xu, C, Tempel, W, Liu, K, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-16 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins.
J.Biol.Chem., 290, 2015
|
|
