8G5M
 
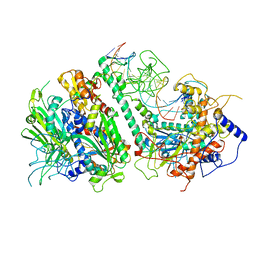 | | Cryo-EM structure of the Mismatch Locking Complex (III) of Human Mitochondrial DNA Polymerase Gamma | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Nayak, A.R, Buchel, G, Herbine, K.H, Sarfallah, A, Sokolova, V.O, Zamudio-Ochoa, A, Temiakov, D. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Structural basis for DNA proofreading.
Nat Commun, 14, 2023
|
|
8G5J
 
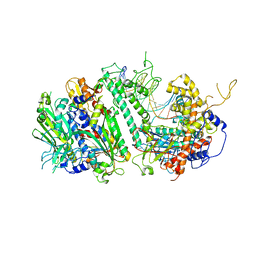 | | Cryo-EM structure of the Mismatch Uncoupling Complex (II) of Human Mitochondrial DNA Polymerase Gamma | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Nayak, A.R, Buchel, G, Herbine, K.H, Sarfallah, A, Sokolova, V.O, Zamudio-Ochoa, A, Temiakov, D. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structural basis for DNA proofreading.
Nat Commun, 14, 2023
|
|
8G5L
 
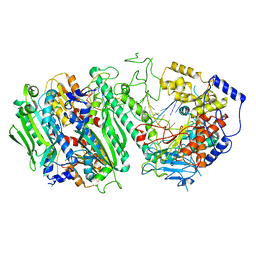 | | Cryo-EM structure of the Primer Separation Complex (IX) of Human Mitochondrial DNA Polymerase Gamma | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Nayak, A.R, Buchel, G, Herbine, K.H, Sarfallah, A, Sokolova, V.O, Zamudio-Ochoa, A, Temiakov, D. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for DNA proofreading.
Nat Commun, 14, 2023
|
|
6XIP
 
 | | The 1.5 A Crystal Structure of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
8G5P
 
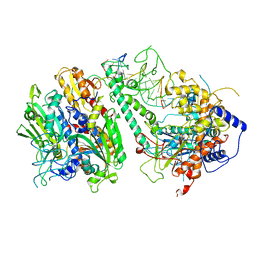 | | Cryo-EM structure of the Guide loop Engagement Complex (V) of Human Mitochondrial DNA Polymerase Gamma | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Nayak, A.R, Buchel, G, Herbine, K.H, Sarfallah, A, Sokolova, V.O, Zamudio-Ochoa, A, Temiakov, D. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structural basis for DNA proofreading.
Nat Commun, 14, 2023
|
|
7PM2
 
 | | Cryo-EM structure of the actomyosin-V complex in the rigor state (central 1er, young JASP-stabilized F-actin, class 4) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-01 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
6ETS
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.333 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
7RU4
 
 | | CC6.33 IgG in complex with SARS-CoV-2-6P-Mut7 S protein (RBD/Fv local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CC6.33 IgG heavy chain Fv, ... | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
8G5O
 
 | | Cryo-EM structure of the Guide loop Engagement Complex (IV) of Human Mitochondrial DNA Polymerase Gamma | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Nayak, A.R, Buchel, G, Herbine, K.H, Sarfallah, A, Sokolova, V.O, Zamudio-Ochoa, A, Temiakov, D. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis for DNA proofreading.
Nat Commun, 14, 2023
|
|
8BTM
 
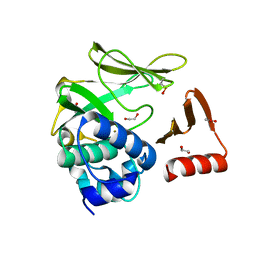 | | Structural and functional studies of geldanamycin amide synthase ShGdmF | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GdmF | | Authors: | Ewert, W, Zeilinger, C, Kirschning, A, Preller, M. | | Deposit date: | 2022-11-29 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and functional studies of geldanamycin amide synthase ShGdmF
To Be Published
|
|
8GJG
 
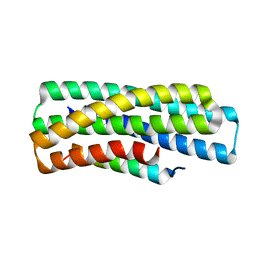 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | gluc_A04_0005, gluc_A04_0005 Binder | | Authors: | Leung, P.J.Y, Bera, A.K, Torres, S.V, Baker, D, Kang, A. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
5LCF
 
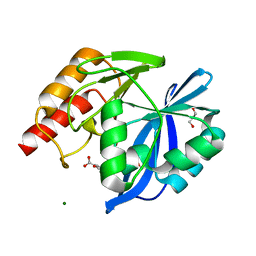 | | VIM-2 metallo-beta-lactamase in complex with 3-oxo-2-phenylisoindoline-4-carboxylic acid (compound 30) | | Descriptor: | 3-oxidanylidene-2-phenyl-1~{H}-isoindole-4-carboxylic acid, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2016-06-21 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | NMR-filtered virtual screening leads to non-metal chelating metallo-beta-lactamase inhibitors.
Chem Sci, 8, 2017
|
|
6F0G
 
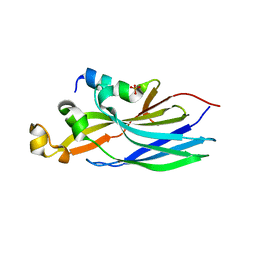 | | Crystal structure ASF1-ip3 | | Descriptor: | Histone chaperone ASF1A, SULFATE ION, ip3 | | Authors: | Gaubert, A, Guichard, B, Richet, N, Le Du, M.H, Andreani, J, Guerois, R, Ochsenbein, F. | | Deposit date: | 2017-11-20 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design on a Rational Basis of High-Affinity Peptides Inhibiting the Histone Chaperone ASF1.
Cell Chem Biol, 26, 2019
|
|
6U9L
 
 | | Imidazole-triggered RAS-specific subtilisin SUBT_BACAM | | Descriptor: | GLYCEROL, POTASSIUM ION, SUBTILISIN BPN', ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J. | | Deposit date: | 2019-09-09 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
7PLV
 
 | | Cryo-EM structure of the actomyosin-V complex in the rigor state (central 1er, class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-01 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PM3
 
 | | Cryo-EM structure of young JASP-stabilized F-actin (central 3er) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-01 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
6OTN
 
 | | Crystal Structure of an N-terminal Fragment of Cancer Associated Tropomyosin 3.1 (Tpm3.1) | | Descriptor: | SULFATE ION, Tropomyosin alpha-3 chain | | Authors: | Rynkiewicz, M.J, Ghosh, A, Lehman, W.J, Janco, M, Gunning, P.W. | | Deposit date: | 2019-05-03 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular integration of the anti-tropomyosin compound ATM-3507 into the coiled coil overlap region of the cancer-associated Tpm3.1.
Sci Rep, 9, 2019
|
|
7PLX
 
 | | Cryo-EM structure of the actomyosin-V complex in the rigor state (central 1er, class 4) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-01 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
6U9S
 
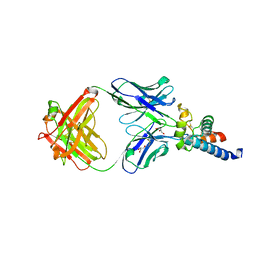 | | Crystal structure of human CD81 large extracellular loop in complex with 5A6 Fab | | Descriptor: | 5A6 FAB Heavy Chain, 5A6 FAB Light Chain, CD81 antigen, ... | | Authors: | Susa, K.J, Seegar, T.C.M, Blacklow, S.C.B, Kruse, A.C. | | Deposit date: | 2019-09-09 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A dynamic interaction between CD19 and the tetraspanin CD81 controls B cell co-receptor trafficking.
Elife, 9, 2020
|
|
5IQS
 
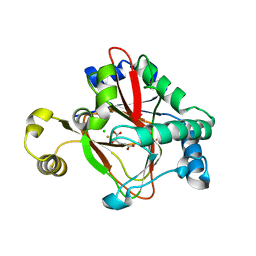 | | WelO5 bound to Fe(II), Cl, and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, FE (II) ION, ... | | Authors: | Mitchell, A.J, Ananth, N, Boal, A.K. | | Deposit date: | 2016-03-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for halogenation by iron- and 2-oxo-glutarate-dependent enzyme WelO5.
Nat.Chem.Biol., 12, 2016
|
|
6OTW
 
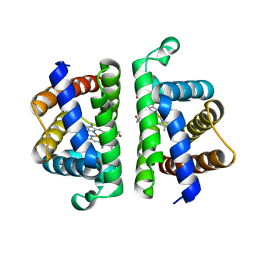 | | Crystallographic Structure of (HbII-HbIII)-O2 from Lucina pectinata at pH 5.0 | | Descriptor: | Hemoglobin II, Hemoglobin III, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Marchany-Rivera, D, Smith, C.A, Rodriguez-Perez, J.D, Lopez-Garriga, J. | | Deposit date: | 2019-05-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | Lucina pectinata oxyhemoglobin (II-III) heterodimer pH susceptibility.
J.Inorg.Biochem., 207, 2020
|
|
6F22
 
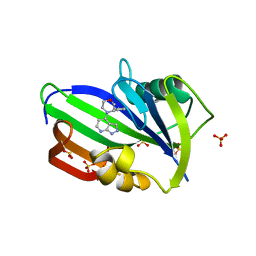 | | Complex between MTH1 and compound 29 (a 4-amino-2,7-diazaindole derivative) | | Descriptor: | (3~{S})-3-phenyl-4-(2~{H}-pyrazolo[3,4-b]pyridin-4-yl)morpholine, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Viklund, J, Talagas, A, Tresaugues, L, Andersson, M, Ericsson, U, Forsblom, R, Ginman, T, Hallberg, K, Lindstrom, J, Persson, L, Silvander, C, Rahm, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design.
J. Med. Chem., 61, 2018
|
|
5LY1
 
 | | JMJD2A/ KDM4A COMPLEXED WITH NI(II) AND Macrocyclic PEPTIDE Inhibitor CP2 (13-mer) | | Descriptor: | CHLORIDE ION, CP2, GLYCEROL, ... | | Authors: | King, O.N.F, Chowdhury, R, Kawamura, A, Schofield, C.J. | | Deposit date: | 2016-09-23 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Highly selective inhibition of histone demethylases by de novo macrocyclic peptides.
Nat Commun, 8, 2017
|
|
6OTY
 
 | | Crystallographic Structure of (HbII-HbIII)-O2 from Lucina pectinata at pH 4.0 | | Descriptor: | Hemoglobin II, Hemoglobin III, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Marchany-Rivera, D, Smith, C.A, Rodriguez-Perez, J.D, Lopez-Garriga, J. | | Deposit date: | 2019-05-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Lucina pectinata oxyhemoglobin (II-III) heterodimer pH susceptibility.
J.Inorg.Biochem., 207, 2020
|
|
7PLT
 
 | | Cryo-EM structure of the actomyosin-V complex in the rigor state (central 1er) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-01 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
