4IMC
 
 | |
5JSI
 
 | | Structure of membrane protein | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Melnikov, I, Polovinkin, V, Kovalev, K, Shevchenko, V, Gushchin, I, Popov, A, Gordeliy, V. | | Deposit date: | 2016-05-08 | | Release date: | 2017-05-31 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fast iodide-SAD phasing for high-throughput membrane protein structure determination.
Sci Adv, 3, 2017
|
|
4ICT
 
 | | Substrate and reaction specificity of Mycobacterium tuberculosis cytochrome P450 CYP121 | | Descriptor: | (3S,6S)-3-benzyl-6-(4-hydroxybenzyl)piperazine-2,5-dione, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fonvielle, M, Le Du, M.-H, Lequin, O, Lecoq, A, Jacquet, M, Thai, R, Dubois, S, Grach, G, Gondry, M, Belin, P. | | Deposit date: | 2012-12-11 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate and Reaction Specificity of Mycobacterium tuberculosis Cytochrome P450 CYP121: INSIGHTS FROM BIOCHEMICAL STUDIES AND CRYSTAL STRUCTURES.
J.Biol.Chem., 288, 2013
|
|
4IDD
 
 | | Structure of the Fragaria x ananassa enone oxidoreductase in complex with NADPH and EHMF | | Descriptor: | (2R)-2-ethyl-4-hydroxy-5-methylfuran-3(2H)-one, 1,2-ETHANEDIOL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the enzymatic formation of the key strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone
J.Biol.Chem., 288, 2013
|
|
3ZSC
 
 | | Catalytic function and substrate recognition of the pectate lyase from Thermotoga maritima | | Descriptor: | 4-deoxy-beta-L-threo-hex-4-enopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid, GLYCEROL, PECTATE TRISACCHARIDE-LYASE, ... | | Authors: | McDonough, M.A, Thymark, M, Frisner, H, Hotchkiss, A, Sonksen, C, Bjornvad, M, Johansen, K.S, Larsen, S. | | Deposit date: | 2011-06-24 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Catalytic Function and Substrate Recognition of the Pectate Lyase from Thermotoga Maritima
To be Published
|
|
6CNK
 
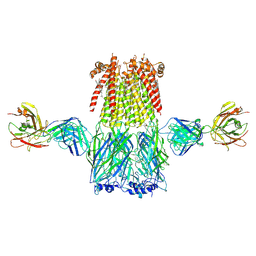 | | Structure of the 3alpha2beta stiochiometry of the human Alpha4Beta2 nicotinic receptor | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Walsh Jr, R.M, Roh, S.H, Gharpure, A, Morales-Perez, C.L, Hibbs, R.E. | | Deposit date: | 2018-03-08 | | Release date: | 2018-05-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural principles of distinct assemblies of the human alpha 4 beta 2 nicotinic receptor.
Nature, 557, 2018
|
|
3ZFZ
 
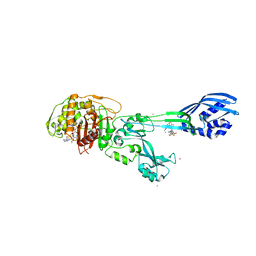 | | Crystal structure of ceftaroline acyl-PBP2a from MRSA with non- covalently bound ceftaroline and muramic acid at allosteric site obtained by soaking | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ceftaroline, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | How Allosteric Control of Staphylococcus Aureus Penicillin Binding Protein 2A Enables Methicillin Resistance and Physiological Function
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6VZU
 
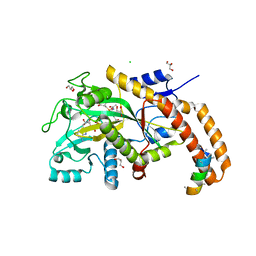 | | TTLL6 bound to alpha-elongation analog | | Descriptor: | (2~{S})-2-[[[(1~{R})-1-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-oxidanyl-phosphoryl]methyl]pentanedioic acid, (2~{S})-2-[[[(1~{R})-1-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.B, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5DF5
 
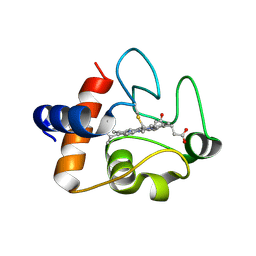 | | The structure of oxidized rat cytochrome c (T28E) at 1.30 angstroms resolution. | | Descriptor: | Cytochrome c, somatic, HEME C, ... | | Authors: | Edwards, B.F.P, Mahapatra, G, Vaishnav, A.A, Brunzelle, J.S, Huttemann, M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | The structure of oxidized rat cytochrome c (T28E) at 1.30 angstroms resolution.
To Be Published
|
|
1JH6
 
 | | Semi-reduced Cyclic Nucleotide Phosphodiesterase from Arabidopsis thaliana | | Descriptor: | SULFATE ION, cyclic phosphodiesterase | | Authors: | Hofmann, A, Grella, M, Botos, I, Filipowicz, W, Wlodawer, A. | | Deposit date: | 2001-06-27 | | Release date: | 2002-02-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the semireduced and inhibitor-bound forms of cyclic nucleotide phosphodiesterase from Arabidopsis thaliana.
J.Biol.Chem., 277, 2002
|
|
2VZT
 
 | | Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with PNP-beta-D-glucosamine | | Descriptor: | 4-nitrophenyl 2-amino-2-deoxy-beta-D-glucopyranoside, ACETATE ION, CADMIUM ION, ... | | Authors: | Lammerts van Bueren, A, Ghinet, M.G, Gregg, K, Fleury, A, Brzezinski, R, Boraston, A.B. | | Deposit date: | 2008-08-05 | | Release date: | 2008-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis of Substrate Recognition in an Exo-Beta-D-Glucosaminidase Involved in Chitosan Hydrolysis.
J.Mol.Biol., 385, 2009
|
|
6PIU
 
 | |
7B7I
 
 | | BAZ2A bromodomain in complex with triazole compound MS04-TN02 | | Descriptor: | 2-[4-[[(~{E})-~{N},~{N}'-dimethylcarbamimidoyl]amino]piperidin-1-yl]-~{N}-[[2-(3-methyl-1,2,3-triazol-4-yl)-1~{H}-benzimidazol-5-yl]methyl]ethanamide, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Sedykh, M, Caflisch, A, Lolli, G. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
5DGO
 
 | |
7B7B
 
 | | BAZ2A bromodomain in complex with triazole compound MS04 | | Descriptor: | 5-ethyl-2-(3-methyl-1,2,3-triazol-4-yl)-1~{H}-benzimidazole, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Sedykh, M, Caflisch, A, Lolli, G. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
8RST
 
 | |
8RSV
 
 | |
1JKX
 
 | | Unexpected formation of an epoxide-derived multisubstrate adduct inhibitor on the active site of GAR transformylase | | Descriptor: | N-[5'-O-PHOSPHONO-RIBOFURANOSYL]-2-[2-HYDROXY-2-[4-[GLUTAMIC ACID]-N-CARBONYLPHENYL]-3-[2-AMINO-4-HYDROXY-QUINAZOLIN-6-YL]-PROPANYLAMINO]-ACETAMIDE, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Greasley, S.E, Marsilje, T.H, Cai, H, Baker, S, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2001-07-13 | | Release date: | 2001-11-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unexpected formation of an epoxide-derived multisubstrate adduct inhibitor on the active site of GAR transformylase.
Biochemistry, 40, 2001
|
|
4UX9
 
 | | Crystal structure of JNK1 bound to a MKK7 docking motif | | Descriptor: | DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 7, MITOGEN-ACTIVATED PROTEIN KINASE 8, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Kragelj, J, Palencia, A, Nanao, M.H, Maurin, D, Bouvignies, G, Blackledge, M, Ringkjobing-Jensen, M. | | Deposit date: | 2014-08-20 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure and Dynamics of the Mkk7-Jnk Signaling Complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1JUH
 
 | | Crystal Structure of Quercetin 2,3-dioxygenase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fusetti, F, Schroeter, K.H, Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2001-08-24 | | Release date: | 2002-05-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the copper-containing quercetin 2,3-dioxygenase from Aspergillus japonicus.
Structure, 10, 2002
|
|
4IC8
 
 | | Crystal structure of the apo ERK5 kinase domain | | Descriptor: | Mitogen-activated protein kinase 7 | | Authors: | Gogl, G, Remenyi, A. | | Deposit date: | 2012-12-10 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanism for the specific assembly and activation of the extracellular signal regulated kinase 5 (ERK5) module.
J.Biol.Chem., 288, 2013
|
|
3ZI8
 
 | | Structure of the R17A mutant of the Ralstonia soleanacerum lectin at 1.5 Angstrom in complex with L-fucose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, MAGNESIUM ION, PUTATIVE FUCOSE-BINDING LECTIN PROTEIN, ... | | Authors: | Arnaud, J, Audfray, A, Claudinon, J, Trondle, K, Trosvalet, M, Thomas, A, Varrot, A, Romer, W, Imberty, A. | | Deposit date: | 2013-01-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reduction of Lectin Valency Drastically Changes Glycolipid Dynamics in Membranes, But not Surface Avidity.
Acs Chem.Biol., 8, 2013
|
|
6PD3
 
 | | Crystal Structure of a H5N1 influenza virus hemagglutinin at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hemagglutinin, ... | | Authors: | Antanasijevic, A, Durst, M.A, Lavie, A, Caffrey, M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a pH sensor in Influenza hemagglutinin using X-ray crystallography.
J.Struct.Biol., 209, 2020
|
|
7ARZ
 
 | | Ternary complex of NAD-dependent formate dehydrogenase from Physcomitrium patens | | Descriptor: | AZIDE ION, Formate dehydrogenase, mitochondrial, ... | | Authors: | Goryaynova, D.A, Nikolaeva, A.Y, Pometun, A.A, Savin, S.S, Parshin, P.D, Popov, V.O, Tishkov, V.I, Boyko, K.M. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ternary complex of NAD-dependent formate dehydrogenase from Physcomitrium patens
To Be Published
|
|
4IDB
 
 | | Structure of the Fragaria x ananassa enone oxidoreductase in complex with NADP+ | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Ripening-induced protein, ... | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the enzymatic formation of the key strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone
J.Biol.Chem., 288, 2013
|
|
