8UBH
 
 | |
7R1U
 
 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the WZ16 inhibitor | | Descriptor: | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
6XZX
 
 | | crystal structure of human carbonic anhydrase I in complex with 4-(3-(2-((2-fluorobenzyl)amino)ethyl)ureido) benzenesulfonamide | | Descriptor: | 1-[2-[(phenylmethyl)amino]ethyl]-3-(3-sulfamoylphenyl)urea, Carbonic anhydrase 1, ZINC ION | | Authors: | Zanotti, G, Majid, A, Bozdag, M, Angeli, A, Carta, F, Berto, P, Supuran, C. | | Deposit date: | 2020-02-05 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Benzylaminoethyureido-Tailed Benzenesulfonamides: Design, Synthesis, Kinetic and X-ray Investigations on Human Carbonic Anhydrases.
Int J Mol Sci, 21, 2020
|
|
6Y00
 
 | | crystal structure of human carbonic anhydrase I in complex with 4-(3-(2-((2-fluorobenzyl)amino)ethyl)ureido) benzenesulfonamide | | Descriptor: | 1-[2-[(4-bromanyl-2-oxidanyl-phenyl)methylamino]ethyl]-3-(3-sulfamoylphenyl)urea, Carbonic anhydrase 1, ZINC ION | | Authors: | Zanotti, G, Majid, A, Bozdag, M, Angeli, A, Carta, F, Berto, P, Supuran, C. | | Deposit date: | 2020-02-05 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Benzylaminoethyureido-Tailed Benzenesulfonamides: Design, Synthesis, Kinetic and X-ray Investigations on Human Carbonic Anhydrases.
Int J Mol Sci, 21, 2020
|
|
7R1T
 
 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the SS148 inhibitor | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
1DJ6
 
 | | COMPLEX OF A Z-DNA HEXAMER, D(CG)3, WITH SYNTHETIC POLYAMINE AT ROOM TEMPERATURE | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*G)-3', MAGNESIUM ION, N,N'-BIS(2-AMINOETHYL)-1,2-ETHANEDIAMINE | | Authors: | Ohishi, H, Tomita, K.-i, Nakanishi, I, Ohtsuchi, M, Hakoshima, T, Rich, A. | | Deposit date: | 1999-12-01 | | Release date: | 1999-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The crystal structure of N1-[2-(2-amino-ethylamino)-ethyl]-ethane-1,2-diamine (polyamines) binding to the minor groove of d(CGCGCG)2, hexamer at room temperature
FEBS Lett., 523, 2002
|
|
5A3E
 
 | |
1DSJ
 
 | | NMR SOLUTION STRUCTURE OF VPR50_75, 20 STRUCTURES | | Descriptor: | VPR PROTEIN | | Authors: | Yao, S, Torres, A.M, Azad, A.A, Macreadie, I.G, Norton, R.S. | | Deposit date: | 1997-10-23 | | Release date: | 1998-07-01 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Helical structure of polypeptides from the C-terminal half of HIV-1 VPR.
Protein Pept.Lett., 5, 1998
|
|
8AII
 
 | | High Resolution Crystal Structure of Enterococcus faecium Nicotinate Nucleotide Adenylyltransferase Complexed with Adenine | | Descriptor: | ADENINE, MAGNESIUM ION, Probable nicotinate-nucleotide adenylyltransferase, ... | | Authors: | Pandian, R, Jeje, O.A, Sayed, Y, Achilonu, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Obtaining high yield recombinant Enterococcus faecium nicotinate nucleotide adenylyltransferase for X-ray crystallography and biophysical studies.
Int.J.Biol.Macromol., 250, 2023
|
|
6YRY
 
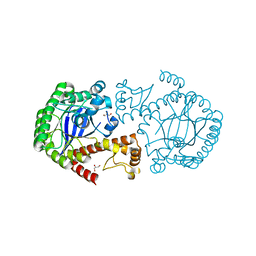 | |
1DEX
 
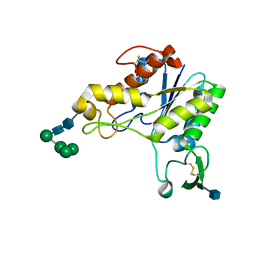 | | RHAMNOGALACTURONAN ACETYLESTERASE FROM ASPERGILLUS ACULEATUS AT 1.9 A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RHAMNOGALACTURONAN ACETYLESTERASE, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Molgaard, A, Kauppinen, S, Larsen, S. | | Deposit date: | 1999-11-16 | | Release date: | 2000-04-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases.
Structure Fold.Des., 8, 2000
|
|
6Y7U
 
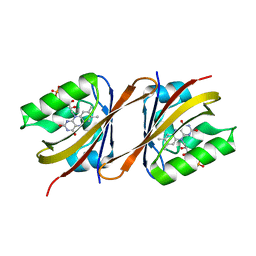 | | Structure of Chloroflexus aggregans Cagg_3753 LOV domain C85A A95P variant (CagFbFP) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gordeliy, V, Gushchin, I. | | Deposit date: | 2020-03-02 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Effects of Proline Substitutions on the Thermostable LOV Domain from Chloroflexus aggregans
Crystals, 2020
|
|
1DKR
 
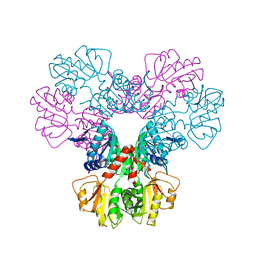 | | CRYSTAL STRUCTURES OF BACILLUS SUBTILIS PHOSPHORIBOSYLPYROPHOSPHATE SYNTHETASE: MOLECULAR BASIS OF ALLOSTERIC INHIBITION AND ACTIVATION. | | Descriptor: | PHOSPHORIBOSYL PYROPHOSPHATE SYNTHETASE, SULFATE ION | | Authors: | Eriksen, T.A, Kadziola, A, Bentsen, A.-K, Harlow, K.W, Larsen, S. | | Deposit date: | 1999-12-08 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the function of Bacillus subtilis phosphoribosyl-pyrophosphate synthetase.
Nat.Struct.Biol., 7, 2000
|
|
6OFW
 
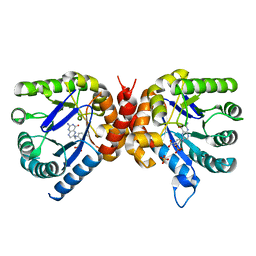 | | Dihydropteroate synthase from Xanthomonas albilineans in complex with 7,8-dihydropteroate | | Descriptor: | 4-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}benzoic acid, Dihydropteroate synthase, SULFATE ION | | Authors: | Oliveira, A.A, Guido, R.V.C, Maluf, F.V, Bueno, R.V, Lima, G.M.A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Dihydropteroate synthase from Xanthomonas albilineans in complex with 7,8-dihydropteroate
To Be Published
|
|
1DLZ
 
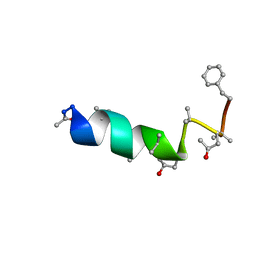 | | SOLUTION STRUCTURE OF THE CHANNEL-FORMER ZERVAMICIN IIB (PEPTAIBOL ANTIBIOTIC) | | Descriptor: | ZERVAMICIN IIB | | Authors: | Balashova, T.A, Shenkarev, Z.O, Tagaev, A.A, Ovchinnikova, T.V, Raap, J, Arseniev, A.S. | | Deposit date: | 1999-12-13 | | Release date: | 2000-02-03 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Channel-Former Zervamicin Iib in Isotropic Solvents.
FEBS Lett., 466, 2000
|
|
4YFG
 
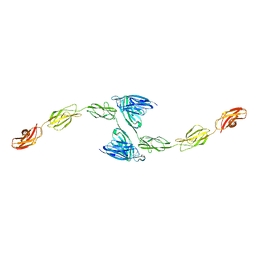 | | Crystal structure of PTP delta meA3/meB minus variant Ig1-Fn1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase delta | | Authors: | Yamagata, A, Fukai, S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.491 Å) | | Cite: | Mechanisms of splicing-dependent trans-synaptic adhesion by PTP delta-IL1RAPL1/IL-1RAcP for synaptic differentiation.
Nat Commun, 6, 2015
|
|
6OQ3
 
 | |
8GAN
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | Cas11, Cas5, Cas7, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8QJT
 
 | | BRM (SMARCA2) Bromodomain in complex with ligand 10 | | Descriptor: | 2-[6-azanyl-5-[(1R,5S)-8-[2-(2-methoxyethoxy)pyridin-4-yl]-3,8-diazabicyclo[3.2.1]octan-3-yl]pyridazin-3-yl]phenol, CHLORIDE ION, Probable global transcription activator SNF2L2, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
5AEK
 
 | | Crystal structure of the human SENP2 C548S in complex with the human SUMO1 K48M F66W | | Descriptor: | SENTRIN-SPECIFIC PROTEASE 2, SMALL UBIQUITIN-RELATED MODIFIER 1 | | Authors: | Gallego, P, Grana-Montes, R, Espargaro, A, Castillo, V, Torrent, J, Lange, R, Papaleo, E, Lindorff-Larsend, K, Ventura, S, Reverter, D. | | Deposit date: | 2014-12-23 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Stepping Back and Forward on Sumo Folding Evolution
To be Published
|
|
8QLP
 
 | | CryoEM structure of the RNA/DNA bound SPARTA (BabAgo/TIR-APAZ) tetrameric complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*AP*TP*TP*AP*GP*AP*GP*CP*CP*GP*TP*C)-3'), MAGNESIUM ION, RNA (5'-R(*AP*UP*GP*AP*CP*GP*GP*CP*UP*CP*UP*AP*AP*UP*CP*UP*AP*UP*UP*AP*GP*U)-3'), ... | | Authors: | Finocchio, G, Koopal, B, Potocnik, A, Heijstek, C, Jinek, M, Swarts, D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Target DNA-dependent activation mechanism of the prokaryotic immune system SPARTA.
Nucleic Acids Res., 52, 2024
|
|
5IXI
 
 | |
7YZS
 
 | |
6OWO
 
 | | CRYO-EM STRUCTURE OF PHOSPHORYLATED AP-2 CORE BOUND TO NECAP | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Partlow, E.A, Baker, R.W, Beacham, G.M, Chappie, J.S, Leschziner, A.E, Hollopeter, G. | | Deposit date: | 2019-05-10 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A structural mechanism for phosphorylation-dependent inactivation of the AP2 complex.
Elife, 8, 2019
|
|
7YZU
 
 | | Crystal structure of the sulfoquinovosyl binding protein SmoF complexed with SQMe | | Descriptor: | Sulfoquinovosyl binding protein, [(2S,3S,4S,5R,6S)-6-methoxy-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Snow, A.J.D, Davies, G.J. | | Deposit date: | 2022-02-21 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The sulfoquinovosyl glycerol binding protein SmoF binds and accommodates plant sulfolipids.
Curr Res Struct Biol, 4, 2022
|
|
