8EIZ
 
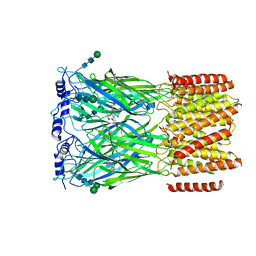 | | Cryo-EM structure of squid sensory receptor CRB1 | | Descriptor: | N-benzyl-2-(2,6-dimethylanilino)-N,N-diethyl-2-oxoethan-1-aminium, Squid sensory receptor CRB1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kang, G, Kim, J.J, Allard, C.A.H, Valencia-Montoya, W.A, van Giesen, L, Kilian, P.B, Bai, X, Bellono, N.W, Hibbs, R.E. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Sensory specializations drive octopus and squid behaviour.
Nature, 616, 2023
|
|
6MHW
 
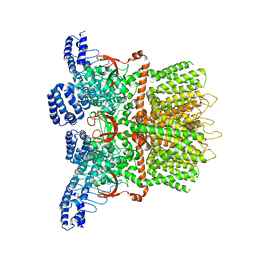 | | Structure of human TRPV3 in the presence of 2-APB in C2 symmetry (1) | | Descriptor: | Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Herzik, M.A, Wu, M, Borschel, W.F, Hirschi, M, Song, A, Lander, G.C, Lee, S.Y. | | Deposit date: | 2018-09-18 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational ensemble of the human TRPV3 ion channel.
Nat Commun, 9, 2018
|
|
7NQE
 
 | |
6MI5
 
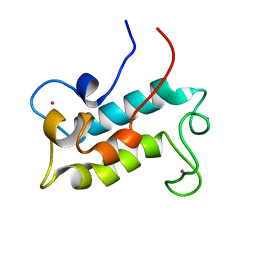 | | NMR solution structure of lanmodulin (LanM) complexed with yttrium(III) ions | | Descriptor: | Lanmodulin, YTTRIUM (III) ION | | Authors: | Cook, E.C, Featherson, E.R, Showalter, S.A, Cotruvo Jr, J.A. | | Deposit date: | 2018-09-19 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Rare Earth Element Recognition by Methylobacterium extorquens Lanmodulin.
Biochemistry, 58, 2019
|
|
7N0N
 
 | |
8EIS
 
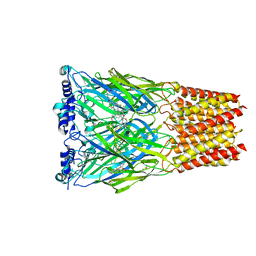 | | Cryo-EM structure of octopus sensory receptor CRT1 | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Octopus sensory receptor | | Authors: | Kang, G, Kim, J.J, Allard, C.A.H, Valencia-Montoya, W.A, Bellono, N.W, Hibbs, R.E. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Sensory specializations drive octopus and squid behaviour.
Nature, 616, 2023
|
|
6RSA
 
 | |
6Y4T
 
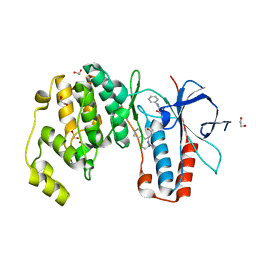 | | Crystal structure of p38 in complex with SR63. | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-1-[[(2~{S})-butan-2-yl]amino]-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
6VA2
 
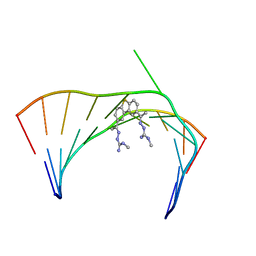 | | Solution Structure of the Tau pre-mRNA Exon 10 Splicing Regulatory Element Bound to MH5 | | Descriptor: | (2E,2'E)-2,2'-{dibenzo[b,d]thiene-2,8-diyldi[(1E)eth-1-yl-1-ylidene]}bis(N-methylhydrazine-1-carboximidamide), RNA (5'-R(*CP*AP*CP*AP*CP*GP*UP*CP*GP*G)-3'), RNA (5'-R(*CP*CP*GP*GP*CP*AP*GP*UP*GP*UP*G)-3') | | Authors: | Chen, J.L, Fountain, M.A, Disney, M.D. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Design, Optimization, and Study of Small Molecules That Target Tau Pre-mRNA and Affect Splicing.
J.Am.Chem.Soc., 142, 2020
|
|
6PBE
 
 | | ZINC17988990-bound TRPV5 in nanodiscs | | Descriptor: | (4-oxo-5-phenyl-3,4-dihydrothieno[2,3-d]pyrimidin-2-yl)methyl 3-(3-oxo-2,3-dihydro-4H-1,4-benzoxazin-4-yl)propanoate, Transient receptor potential cation channel subfamily V member 5 | | Authors: | Hughes, T.E.T, Rosario, J.S.D, Kapoor, A, Yazici, A.T, Fluck, E.C, Filizola, M, Rohacs, T, Moiseenkova-Bell, V.Y. | | Deposit date: | 2019-06-13 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structure-based characterization of novel TRPV5 inhibitors.
Elife, 8, 2019
|
|
5K9J
 
 | | Crystal structure of multidonor HV6-1-class broadly neutralizing Influenza A antibody 56.a.09 isolated following H5 immunization. | | Descriptor: | 56.a.09 heavy chain, 56.a.09 light chain, POLYETHYLENE GLYCOL (N=34) | | Authors: | Joyce, M.G, Thomas, P.V, Wheatley, A.K, McDermott, A.B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-31 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Cell, 166, 2016
|
|
5M9F
 
 | |
6MP8
 
 | | X-ray crystal structure of VioC bound to Fe(II), D-arginine, and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent L-arginine hydroxylase, D-ARGININE, ... | | Authors: | Dunham, N.P, Boal, A.K. | | Deposit date: | 2018-10-05 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | alpha-Amine Desaturation of d-Arginine by the Iron(II)- and 2-(Oxo)glutarate-Dependent l-Arginine 3-Hydroxylase, VioC.
Biochemistry, 57, 2018
|
|
8C8V
 
 | | Priestia megaterium mupirocin-resistant isoleucyl-tRNA synthetase 2 with a fully-resolved C-terminal tRNA-binding domain complexed with an isoleucyl-adenylate analogue | | Descriptor: | D(-)-TARTARIC ACID, Isoleucine--tRNA ligase, N-[ISOLEUCINYL]-N'-[ADENOSYL]-DIAMINOSUFONE, ... | | Authors: | Brkic, A, Leibundgut, M, Jablonska, J, Zanki, V, Car, Z, Petrovic Perokovic, V, Ban, N, Gruic-Sovulj, I. | | Deposit date: | 2023-01-21 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Nat Commun, 14, 2023
|
|
6Y3R
 
 | | 14-3-3 Sigma in complex with phosphorylated (Thr391) Gab2 peptide | | Descriptor: | 14-3-3 protein sigma, Chain P, MAGNESIUM ION | | Authors: | Ballone, A, Lau, R.A, Zweipfenning, F.P.A, Ottmann, C. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A new soaking procedure for X-ray crystallographic structural determination of protein-peptide complexes.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7AOS
 
 | | crystal structure of the RARalpha/RXRalpha ligand binding domain heterodimer in complex with a fragment of SRC1 coactivator | | Descriptor: | 4-{[(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)carbonyl]amino}benzoic acid, 6-[1-(3,5,5,8,8-PENTAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)CYCLOPROPYL]PYRIDINE-3-CARBOXYLIC ACID, GLYCEROL, ... | | Authors: | le Maire, A, Guee, L, Bourguet, W. | | Deposit date: | 2020-10-15 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insights into the Interaction of the Intrinsically Disordered Co-activator TIF2 with Retinoic Acid Receptor Heterodimer (RXR/RAR).
J.Mol.Biol., 433, 2021
|
|
7Q3H
 
 | | Pentameric ligand-gated ion channel, DeCLIC at pH 7 with 10 mM EDTA | | Descriptor: | Neur_chan_LBD domain-containing protein | | Authors: | Lycksell, M, Rovsnik, U, Hanke, A, Howard, R.J, Lindahl, E. | | Deposit date: | 2021-10-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Biophysical characterization of calcium-binding and modulatory-domain dynamics in a pentameric ligand-gated ion channel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ATV
 
 | | Structure of protein kinase ck2 catalytic subunit (csnk2a2 gene product) in complex with the bivalent inhibitor KN2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Casein kinase II subunit alpha', ... | | Authors: | Lindenblatt, D, Applegate, V, Nickelsen, A, Klussmann, M, Neundorf, I, Goetz, C, Jose, J, Niefind, K. | | Deposit date: | 2020-10-31 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Molecular Plasticity of Crystalline CK2 alpha ' Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity.
J.Med.Chem., 65, 2022
|
|
8CPB
 
 | | 1,6-anhydro-n-actetylmuramic acid kinase (AnmK) in complex with AMPPNP, and AnhMurNAc at 1.7 Angstroms resolution. | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, Anhydro-N-acetylmuramic acid kinase, GLYCEROL, ... | | Authors: | Jimenez-Faraco, E, Hermoso, J.A. | | Deposit date: | 2023-03-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic process of anhydro-N-acetylmuramic acid kinase from Pseudomonas aeruginosa.
J.Biol.Chem., 299, 2023
|
|
6N5E
 
 | |
8CP9
 
 | |
5MF4
 
 | | Tubulin-Dictyostatin complex | | Descriptor: | (3~{Z},5~{E},7~{R},8~{S},10~{S},11~{Z},13~{S},14~{R},15~{S},17~{S},20~{R},21~{S},22~{S})-22-[(2~{S},3~{Z})-hexa-3,5-dien-2-yl]-7,13,15,17,21-pentamethyl-8,10,14,20-tetrakis(oxidanyl)-1-oxacyclodocosa-3,5,11-trien-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Trigili, C, Barasoain, I, Sanchez-Murcia, P.A, Bargsten, K, Redondo-Horcajo, M, Nogales, A, Gardner, N.M, Meyer, A, Naylor, G.J, Gomez-Rubio, E, Gago, F, Steinmetz, M.O, Paterson, I, Prota, A.E, Diaz, J.F. | | Deposit date: | 2016-11-17 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Determinants of the Dictyostatin Chemotype for Tubulin Binding Affinity and Antitumor Activity Against Taxane- and Epothilone-Resistant Cancer Cells.
ACS Omega, 1, 2016
|
|
7N0A
 
 | | Structure of Human Leukaemia Inhibitory Factor with Fab MSC1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Leukemia inhibitory factor, ... | | Authors: | Raman, S, Bosch, A, Fransson, J, Julien, J.P. | | Deposit date: | 2021-05-25 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Therapeutic Targeting of LIF Overcomes Macrophage-mediated Immunosuppression of the Local Tumor Microenvironment.
Clin.Cancer Res., 29, 2023
|
|
6PHJ
 
 | |
6PHO
 
 | |
