8V9Q
 
 | | Crystal structure of mGalNAc-T1 in complex with the mucin glycopeptide Muc5AC-13, Mn2+, and UDP. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Samara, N.L, Collette, A.M. | | Deposit date: | 2023-12-08 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | An unusual dual sugar-binding lectin domain controls the substrate specificity of a mucin-type O-glycosyltransferase.
Sci Adv, 10, 2024
|
|
7O31
 
 | | Crystal structure of the anti-PAS Fab 1.2 in complex with its epitope peptide and the anti-Kappa VHH domain | | Descriptor: | 1,2-ETHANEDIOL, PAS#1 epitope peptide, anti-Kappa VHH domain, ... | | Authors: | Schilz, J, Schiefner, A, Skerra, A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular recognition of structurally disordered Pro/Ala-rich sequences (PAS) by antibodies involves an Ala residue at the hot spot of the epitope.
J.Mol.Biol., 433, 2021
|
|
7RAH
 
 | | Adenylate cyclase toxin RTX domain fragment bound to M1H5 Fab and M2B10 Fab | | Descriptor: | Bifunctional adenylate cyclase toxin/hemolysin CyaA,Bifunctional adenylate cyclase toxin/hemolysin CyaA, CALCIUM ION, M1H5 Fab Heavy Chain, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2021-07-01 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for antibody binding to adenylate cyclase toxin reveals RTX linkers as neutralization-sensitive epitopes.
Plos Pathog., 17, 2021
|
|
6Q31
 
 | | Dye type peroxidase Aa from Streptomyces lividans: 156.8 kGy structure | | Descriptor: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ebrahim, A, Moreno-Chicano, T, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Appleby, M, Owen, R.L. | | Deposit date: | 2018-12-03 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins.
Iucrj, 6, 2019
|
|
7SSO
 
 | |
8OIP
 
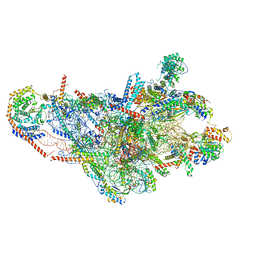 | | 28S mammalian mitochondrial small ribosomal subunit with mtRF1 and P-site tRNA | | Descriptor: | 12S rRNA, 28S ribosomal protein S15, mitochondrial, ... | | Authors: | Saurer, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Ban, N. | | Deposit date: | 2023-03-23 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of translation termination at noncanonical stop codons in human mitochondria.
Science, 380, 2023
|
|
7NT6
 
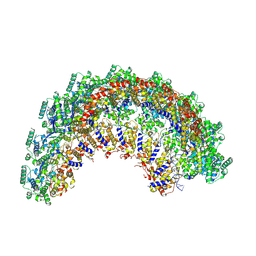 | | CryoEM structure of the Nipah virus nucleocapsid spiral clam-shaped assembly | | Descriptor: | Nucleoprotein, RNA (42-MER), RNA (48-MER) | | Authors: | Ker, D.S, Jenkins, H.T, Greive, S.J, Antson, A.A. | | Deposit date: | 2021-03-09 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | CryoEM structure of the Nipah virus nucleocapsid assembly.
Plos Pathog., 17, 2021
|
|
6Q5A
 
 | |
5LDZ
 
 | | Quadruple space group ambiguity due to rotational and translational non-crystallographic symmetry in human liver fructose-1,6-bisphosphatase | | Descriptor: | CHLORIDE ION, Fructose-1,6-bisphosphatase 1, SULFATE ION, ... | | Authors: | Ruf, A, Tetaz, T, Schott, B, Joseph, C, Rudolph, M.G. | | Deposit date: | 2016-06-29 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Quadruple space-group ambiguity owing to rotational and translational noncrystallographic symmetry in human liver fructose-1,6-bisphosphatase.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8G8W
 
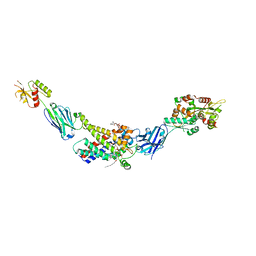 | | Molecular mechanism of nucleotide inhibition of human uncoupling protein 1 | | Descriptor: | CARDIOLIPIN, GUANOSINE-5'-TRIPHOSPHATE, Mitochondrial brown fat uncoupling protein 1, ... | | Authors: | Gogoi, P, Jones, S.A, Ruprecht, J.J, King, M.S, Lee, Y, Zogg, T, Pardon, E, Chand, D, Steimle, S, Copeman, D, Cotrim, C.A, Steyaert, J, Crichton, P.G, Moiseenkova-Bell, V, Kunji, E.R.S. | | Deposit date: | 2023-02-20 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of purine nucleotide inhibition of human uncoupling protein 1.
Sci Adv, 9, 2023
|
|
5LGA
 
 | | Structural analysis and biological activities of BXL0124, a Gemini analog of Vitamin D | | Descriptor: | (1~{R},3~{S},5~{Z})-5-[2-[(1~{R},3~{a}~{S},7~{a}~{R})-7~{a}-methyl-1-[(6~{R})-1,1,1-tris(fluoranyl)-10-methyl-2,10-bis(oxidanyl)-2-(trifluoromethyl)undeca-3,8-diyn-6-yl]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, SRC-2, Vitamin D3 receptor A | | Authors: | Belorusova, A.Y, Rochel, N. | | Deposit date: | 2016-07-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis and biological activities of BXL0124, a gemini analog of vitamin D.
J. Steroid Biochem. Mol. Biol., 173, 2017
|
|
7O33
 
 | | Crystal structure of the anti-PAS Fab 3.1 in complex with its epitope peptide | | Descriptor: | APSA epitope peptide, anti-PAS Fab 3.1 chimeric heavy chain, anti-PAS Fab 3.1 chimeric light chain | | Authors: | Schilz, J, Skerra, A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of structurally disordered Pro/Ala-rich sequences (PAS) by antibodies involves an Ala residue at the hot spot of the epitope.
J.Mol.Biol., 433, 2021
|
|
7NT5
 
 | | CryoEM structure of the Nipah virus nucleocapsid single helical turn assembly | | Descriptor: | Nucleoprotein, RNA (78-MER) | | Authors: | Ker, D.S, Jenkins, H.T, Greive, S.J, Antson, A.A. | | Deposit date: | 2021-03-09 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | CryoEM structure of the Nipah virus nucleocapsid assembly.
Plos Pathog., 17, 2021
|
|
6Q20
 
 | |
6Q34
 
 | | Dye type peroxidase Aa from Streptomyces lividans: 196 kGy structure | | Descriptor: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ebrahim, A, Moreno-Chicano, T, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Appleby, M, Owen, R.L. | | Deposit date: | 2018-12-03 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins.
Iucrj, 6, 2019
|
|
6WCK
 
 | | KRAS G-quadruplex G16T mutant with Bromo Uracil replacing T8 and T16. | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*CP*GP*GP*(BRU)P*GP*TP*GP*GP*GP*AP*AP*(BRU)P*AP*GP*GP*GP*AP*A)-3'), POTASSIUM ION | | Authors: | Schmidberger, J.W, Ou, A, Smith, N.M, Iyer, K.S, Bond, C.S. | | Deposit date: | 2020-03-30 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | High resolution crystal structure of a KRAS promoter G-quadruplex reveals a dimer with extensive poly-A pi-stacking interactions for small-molecule recognition.
Nucleic Acids Res., 48, 2020
|
|
6Q3M
 
 | | Structure of CHD4 PHD2 - tandem chromodomains | | Descriptor: | 1,2-ETHANEDIOL, Chromodomain-helicase-DNA-binding protein 4, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Alt, A, Mancini, E.J. | | Deposit date: | 2018-12-04 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of histone readers
To Be Published
|
|
8P2K
 
 | | Ternary complex of translating ribosome, NAC and METAP1 | | Descriptor: | 18s rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Jia, M, Jaskolowski, M, Scaiola, A, Jomaa, A, Ban, N. | | Deposit date: | 2023-05-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | NAC controls cotranslational N-terminal methionine excision in eukaryotes.
Science, 380, 2023
|
|
6YL7
 
 | |
7OBQ
 
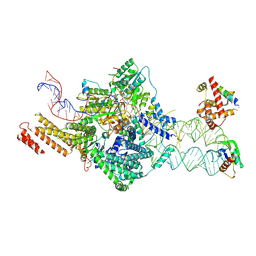 | | SRP-SR at the distal site conformation | | Descriptor: | EM14S01-3B_G0054400.mRNA.1.CDS.1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jomaa, A, Ban, N. | | Deposit date: | 2021-04-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular mechanism of cargo recognition and handover by the mammalian signal recognition particle.
Cell Rep, 36, 2021
|
|
6PUB
 
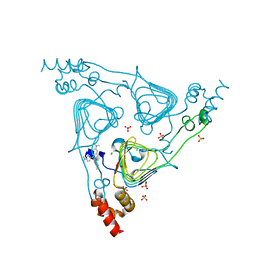 | | Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae in the Complex with Crystal Violet | | Descriptor: | CHLORIDE ION, CRYSTAL VIOLET, Chloramphenicol acetyltransferase, ... | | Authors: | Kim, Y, Maltseva, N, Kuhn, M, Stam, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae in the Complex with Crystal Violet
To Be Published
|
|
8OO2
 
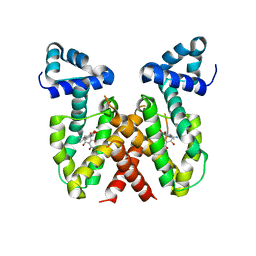 | | ChdA complex with amido-chelocardin | | Descriptor: | 2-carboxamido-2-deacetyl-chelocardin, MAGNESIUM ION, Putative transcriptional regulator | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2023-04-04 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Revision of the Absolute Configurations of Chelocardin and Amidochelocardin.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7OBR
 
 | | RNC-SRP early complex | | Descriptor: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Ban, N. | | Deposit date: | 2021-04-23 | | Release date: | 2021-07-21 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular mechanism of cargo recognition and handover by the mammalian signal recognition particle.
Cell Rep, 36, 2021
|
|
8G7F
 
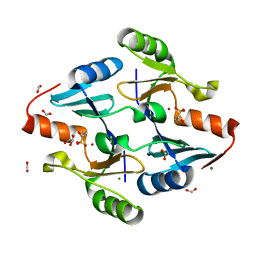 | | Crystal Structure of FosB from Bacillus cereus with Zinc and 1-hydroxypropylphosphonic acid | | Descriptor: | FORMIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Travis, S, Pang, A.H, Tsodikov, O.V, Garneau-Tsodikova, S, Thompson, M.K. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Identification and analysis of small molecule inhibitors of FosB from Staphylococcus aureus.
Rsc Med Chem, 14, 2023
|
|
5MQM
 
 | | Glycoside hydrolase BT_0986 | | Descriptor: | CALCIUM ION, D-rhamnopyranose tetrazole, Glycosyl hydrolases family 2, ... | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
